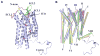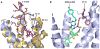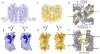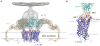Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists - PubMed (original) (raw)
. 2010 Nov 19;330(6007):1066-71.
doi: 10.1126/science.1194396. Epub 2010 Oct 7.
Ellen Y T Chien, Clifford D Mol, Gustavo Fenalti, Wei Liu, Vsevolod Katritch, Ruben Abagyan, Alexei Brooun, Peter Wells, F Christopher Bi, Damon J Hamel, Peter Kuhn, Tracy M Handel, Vadim Cherezov, Raymond C Stevens
Affiliations
- PMID: 20929726
- PMCID: PMC3074590
- DOI: 10.1126/science.1194396
Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists
Beili Wu et al. Science. 2010.
Abstract
Chemokine receptors are critical regulators of cell migration in the context of immune surveillance, inflammation, and development. The G protein-coupled chemokine receptor CXCR4 is specifically implicated in cancer metastasis and HIV-1 infection. Here we report five independent crystal structures of CXCR4 bound to an antagonist small molecule IT1t and a cyclic peptide CVX15 at 2.5 to 3.2 angstrom resolution. All structures reveal a consistent homodimer with an interface including helices V and VI that may be involved in regulating signaling. The location and shape of the ligand-binding sites differ from other G protein-coupled receptors and are closer to the extracellular surface. These structures provide new clues about the interactions between CXCR4 and its natural ligand CXCL12, and with the HIV-1 glycoprotein gp120.
Figures
Fig. 1
Overall fold of the CXCR4/IT1t complex and comparison with other GPCR structures. (A) Overall fold of the CXCR4-2/IT1t. The receptor is colored blue. The N-terminus, ECL1, ECL2 and ECL3 are highlighted in brown, blue, green and red, respectively. The compound IT1t is shown in a magenta stick representation. The disulfide bonds are yellow. Conserved water molecules (67) are shown as red spheres. (B) Comparison of TM helices for CXCR4 (blue), β2AR (PDB ID: 2RH1; yellow), A2AAR (PDB ID: 3EML; green) and rhodopsin (PDB ID: 1U19; pink).
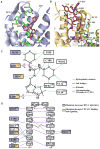
Fig. 2
CXCR4 ligand-binding cavities for the small molecule IT1t and the cyclic peptide CVX15. (A) CXCR4 ligand-binding cavity for the small molecule IT1t. IT1t (magenta) and the residues of the receptor (green) involved in the ligand interactions are shown in stick representation. Nitrogen atoms are blue and sulfur atoms are yellow. Hydrophobic contacts are shown as green dashed lines, salt bridges are red, and hydrogen bonds are blue. Only the helices involved in the receptor-ligand interaction and part of ECL2 are shown. (B) CXCR4 ligand-binding cavity for the peptide CVX15. The residues of CVX15 (brown) and the residues of the receptor (green) involved in receptor-ligand polar interactions are shown in stick representation. The Cys4-Cys13 disulfide bridge in CVX15 is shown as a yellow stick. Intramolecular hydrogen bonds of CVX15 are shown as purple dashed lines. (C) Schematic representation of the interactions between CXCR4 and IT1t in the ligand binding pocket. Mutations reported to decrease HIV-1 infectivity and disrupt CXCL12 binding and signaling are indicated with blue and yellow squares, respectively (56, 68). (D) Schematic representation of the interactions between CXCR4 and CVX15 in the ligand binding pocket.
Fig. 3
CXCR4 ligand-binding modes and comparison with other GPCR structures. (A) Comparison of the ligand-binding modes for IT1t and CVX15. CXCR4 molecules in the CXCR4- 2/IT1t and CXCR4-3/CVX15 complexes are colored blue and yellow, respectively. IT1t (magenta) and CVX15 (brown) are shown as sticks. (B) Comparison of the small molecule ligand-binding modes for CXCR4, β2AR (PDB ID: 2RH1), A2AAR (PDB ID: 3EML) and rhodopsin (PDB ID: 1U19). Only CXCR4 helices are shown (blue). The ligands IT1t (for CXCR4, magenta), carazolol (for β2AR, yellow), ZM241385 (for A2AAR, cyan) and retinal (for rhodopsin, green) are shown in stick representation.
Fig. 4
Dimer interactions in CXCR4-2/IT1t and CXCR4-3/CVX15. (A) Molecular surface representation of the CXCR4 dimer in CXCR4-2/IT1t (blue). (B) Dimer interface in CXCR4-2/IT1t. The surface involved in dimerization is highlighted in dark blue. (C) Molecular surface representation of the CXCR4 dimer in CXCR4-3/CVX15 (yellow). A hypothetical path of the C-terminus, which is not observed in the CXCR4-3/CVX15 structure, is shown as a dashed curve. (D) Dimer interface in CXCR4-3/CVX15. The surface involved in dimer interaction is highlighted in orange. (E) Top view of the extracellular side of the dimers. Two structures show similar interactions via helices V and VI. Residues of CXCR4-2/IT1t involved in the dimer interaction are shown in stick representation, and colored blue in molecule A, cyan in molecule B. (F) Bottom view of the intracellular side of the dimers. Contacts can only be observed at the intracellular tips of helices III and IV, and ICL2 in CXCR4-3/CVX15. The residues of CXCR4-3/CVX15 involved in the dimer interaction are shown in stick representation, and colored yellow and orange. These interactions are not present in the CXCR4/IT1t complex.
Fig. 5
Stoichiometry of possible CXCR4/CXCL12 binding/signaling complexes. No information on the orientation of CXCL12 with respect to CXCR4 is implied from the models presented. (A) Monomeric CXCR4 binding monomeric CXCL12, (B) dimeric CXCR4 binding monomeric CXCL12, (C) dimeric CXCR4 binding dimeric CXCL12 at either one or both orthosteric sites on each protomer. Alternatively, the 2:2 complex could involve two CXCL12 monomers binding dimeric CXCR4 (not shown). Both CXCR4 and CXCL12 surfaces are colored according to their electrostatic potential from red (negative) to blue (positive), highlighting the charge complementarity of these proteins. The portion of the CXCR4 N-terminal domain (CXCR4-N) present in both the CXCL12 complex (PDB ID: 2K05) and crystal structures of this study is colored yellow, while the remainder is purple (Site 1). Pro27 and the three sulfotyrosines from the CXCR4 N-terminus are represented with space-filling models. The CVX15 peptide (green ribbon) is shown in one CXCR4 receptor per panel and suggests the binding site for Lys1 and the rest of the flexible N-terminal region of CXCL12, which is critical for receptor activation (Site 2). Figures were prepared using ICM software (
).
Fig. 6
Model of early stages of the HIV-1 entry process. (A) Viral entry begins with binding of envelope spikes consisting of a heterotrimer (gp120)3 (gp41)3 (gray wire, EM databank ID: emd_5020 and emd_5023; PDB ID: 3DNO) to CD4 on the surface of host target cells. Glycoprotein gp120 (core structure, cyan, PDB ID: 2QAD) interacts with CD4 (tan, PDB ID: 1WIP and 2KLU). This interaction triggers conformational changes in gp120 that increase the exposure of the third variable loop V3 (magenta) and a region of gp120 between inner and outer domains. CCR5 or CXCR4 (blue) is then recruited as a co-receptor. The number of spikes involved in viral entry and the number of molecules of CD4 or CXCR4 binding to a single spike are unknown; here three CD4 molecules are represented, which results in the close approach of gp120 molecules to the host cell membrane where the interaction with three CXCR4 molecules is depicted. (B) By analogy to a two-site model based on CCR5 (64), the N-terminus of CXCR4 containing sulfotyrosines (site1, circled in yellow) binds first to the base of the V3 loop inducing further conformational changes in gp120 that enable V3 to bind to the extracellular side of CXCR4, primarily ECL2, ECL3 and the ligand-binding cavity (site 2, circled in yellow). CXCR4 residues previously shown to affect gp120 binding are shown as sticks with carbons colored in orange. A hypothetical path of the CXCR4 N-terminus, which is not observed in the current structure, is shown as a blue dashed curve. Only CXCR4 monomers are shown for clarity, although dimers are also possible. Fig. 1, 2, 3, 4 and 6 were prepared using PyMOL.
Comment in
- G protein-coupled receptors: Insights into chemokine receptors.
Harrison C. Harrison C. Nat Rev Drug Discov. 2010 Dec;9(12):920. doi: 10.1038/nrd3331. Nat Rev Drug Discov. 2010. PMID: 21119730 No abstract available.
Similar articles
- Anti-HIV small-molecule binding in the peptide subpocket of the CXCR4:CVX15 crystal structure.
Cox BD, Prosser AR, Katzman BM, Alcaraz AA, Liotta DC, Wilson LJ, Snyder JP. Cox BD, et al. Chembiochem. 2014 Jul 21;15(11):1614-20. doi: 10.1002/cbic.201402056. Epub 2014 Jul 2. Chembiochem. 2014. PMID: 24990206 Free PMC article. - Chemokine receptor CXCR4 oligomerization is disrupted selectively by the antagonist ligand IT1t.
Ward RJ, Pediani JD, Marsango S, Jolly R, Stoneman MR, Biener G, Handel TM, Raicu V, Milligan G. Ward RJ, et al. J Biol Chem. 2021 Jan-Jun;296:100139. doi: 10.1074/jbc.RA120.016612. Epub 2020 Dec 6. J Biol Chem. 2021. PMID: 33268380 Free PMC article. - Progress toward rationally designed small-molecule peptide and peptidomimetic CXCR4 antagonists.
Våbenø J, Haug BE, Rosenkilde MM. Våbenø J, et al. Future Med Chem. 2015;7(10):1261-83. doi: 10.4155/fmc.15.64. Future Med Chem. 2015. PMID: 26144264 Review. - Orally bioavailable isothioureas block function of the chemokine receptor CXCR4 in vitro and in vivo.
Thoma G, Streiff MB, Kovarik J, Glickman F, Wagner T, Beerli C, Zerwes HG. Thoma G, et al. J Med Chem. 2008 Dec 25;51(24):7915-20. doi: 10.1021/jm801065q. J Med Chem. 2008. PMID: 19053768 - Structure-based studies of chemokine receptors.
Zhu L, Zhao Q, Wu B. Zhu L, et al. Curr Opin Struct Biol. 2013 Aug;23(4):539-46. doi: 10.1016/j.sbi.2013.05.003. Epub 2013 May 22. Curr Opin Struct Biol. 2013. PMID: 23706951 Review.
Cited by
- Chemokinergic and Dopaminergic Signalling Collaborates through the Heteromer Formed by CCR9 and Dopamine Receptor D5 Increasing the Migratory Speed of Effector CD4+ T-Cells to Infiltrate the Colonic Mucosa.
Campos J, Osorio-Barrios F, Villanelo F, Gutierrez-Maldonado SE, Vargas P, Pérez-Acle T, Pacheco R. Campos J, et al. Int J Mol Sci. 2024 Sep 18;25(18):10022. doi: 10.3390/ijms251810022. Int J Mol Sci. 2024. PMID: 39337509 Free PMC article. - Structural insights into CXCR4 modulation and oligomerization.
Saotome K, McGoldrick LL, Ho JH, Ramlall TF, Shah S, Moore MJ, Kim JH, Leidich R, Olson WC, Franklin MC. Saotome K, et al. Nat Struct Mol Biol. 2024 Sep 23. doi: 10.1038/s41594-024-01397-1. Online ahead of print. Nat Struct Mol Biol. 2024. PMID: 39313635 - Applying Molecular Modeling to the Design of Innovative, Non-Symmetrical CXCR4 Inhibitors with Potent Anticancer Activity.
Martínez-Asensio M, Sàrrias L, Gorjón-de-Pablo G, Fernández-Serrano M, Camaló-Vila J, Gibert A, Puig de la Bellacasa R, Teixidó J, Roué G, Borrell JI, Estrada-Tejedor R. Martínez-Asensio M, et al. Int J Mol Sci. 2024 Aug 30;25(17):9446. doi: 10.3390/ijms25179446. Int J Mol Sci. 2024. PMID: 39273392 Free PMC article. - Interaction and dynamics of chemokine receptor CXCR4 binding with CXCL12 and hBD-3.
Penfield J, Zhang L. Penfield J, et al. Commun Chem. 2024 Sep 13;7(1):205. doi: 10.1038/s42004-024-01280-6. Commun Chem. 2024. PMID: 39271963 Free PMC article. - Allosteric modulation of the CXCR4:CXCL12 axis by targeting receptor nanoclustering via the TMV-TMVI domain.
García-Cuesta EM, Martínez P, Selvaraju K, Ulltjärn G, Gómez Pozo AM, D'Agostino G, Gardeta S, Quijada-Freire A, Blanco Gabella P, Roca C, Hoyo DD, Jiménez-Saiz R, García-Rubia A, Soler Palacios B, Lucas P, Ayala-Bueno R, Santander Acerete N, Carrasco Y, Oscar Sorzano C, Martinez A, Campillo NE, Jensen LD, Rodriguez Frade JM, Santiago C, Mellado M. García-Cuesta EM, et al. Elife. 2024 Sep 9;13:RP93968. doi: 10.7554/eLife.93968. Elife. 2024. PMID: 39248648 Free PMC article.
References
- Baggiolini M. Nature. 1998;392:565. - PubMed
- Moser B, Wolf M, Walz A, Loetscher P. Trends Immunol. 2004;25:75. - PubMed
- Mackay CR. Nat Immunol. 2001;2:95. - PubMed
- Zou YR, Kottmann AH, Kuroda M, Taniuchi I, Littman DR. Nature. 1998;393:595. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R21 RR025336-01A1/RR/NCRR NIH HHS/United States
- R21 RR025336/RR/NCRR NIH HHS/United States
- P50 GM073197-07/GM/NIGMS NIH HHS/United States
- F32 GM083463-03/GM/NIGMS NIH HHS/United States
- R21 AI087189/AI/NIAID NIH HHS/United States
- GM075915/GM/NIGMS NIH HHS/United States
- R01 GM075915/GM/NIGMS NIH HHS/United States
- Y1-GM-1104/GM/NIGMS NIH HHS/United States
- R21 AI087189-02/AI/NIAID NIH HHS/United States
- F32 GM083463/GM/NIGMS NIH HHS/United States
- R01 GM081763/GM/NIGMS NIH HHS/United States
- R01 AI037113-13/AI/NIAID NIH HHS/United States
- P50 GM073197/GM/NIGMS NIH HHS/United States
- U54 GM094618/GM/NIGMS NIH HHS/United States
- R01 AI037113/AI/NIAID NIH HHS/United States
- R01 GM081763-03/GM/NIGMS NIH HHS/United States
- U54 GM074961-050001/GM/NIGMS NIH HHS/United States
- R01 GM089857/GM/NIGMS NIH HHS/United States
- Y1-CO-1020/CO/NCI NIH HHS/United States
- R01 GM071872/GM/NIGMS NIH HHS/United States
- U54 GM074961/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases