genenames.org: the HGNC resources in 2011 - PubMed (original) (raw)
. 2011 Jan;39(Database issue):D514-9.
doi: 10.1093/nar/gkq892. Epub 2010 Oct 6.
Affiliations
- PMID: 20929869
- PMCID: PMC3013772
- DOI: 10.1093/nar/gkq892
genenames.org: the HGNC resources in 2011
Ruth L Seal et al. Nucleic Acids Res. 2011 Jan.
Abstract
The HUGO Gene Nomenclature Committee (HGNC) aims to assign a unique gene symbol and name to every human gene. The HGNC database currently contains almost 30,000 approved gene symbols, over 19,000 of which represent protein-coding genes. The public website, www.genenames.org, displays all approved nomenclature within Symbol Reports that contain data curated by HGNC editors and links to related genomic, phenotypic and proteomic information. Here we describe improvements to our resources, including a new Quick Gene Search, a new List Search, an integrated HGNC BioMart and a new Statistics and Downloads facility.
Figures
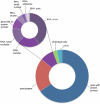
Figure 1.
The proportion of HGNC gene symbols annotated with each locus type. The main doughnut chart shows the proportions of major locus groups. The purple region represents genes annotated with the non-protein-coding RNA locus group; the smaller chart shows the proportion of RNA genes annotated with each RNA-specific locus type. The ‘RNA, misc’ category groups together RNA locus types that represent a small number of genes. These are: ‘RNA, Cajal body-specific’, ‘RNA, ribosomal’, ‘RNA, small cytoplasmic’, ‘RNA, vault’, ‘RNA, Y’, ‘RNA, RNase’ and ‘RNA, telomerase’. A full list of locus types, along with total numbers of approved symbols for each category, can be viewed at our Statistics and Downloads facility (
).
Figure 2.
An example of the results output generated by the HGNC List Search. The ‘Match Type’ column shows how each submitted symbol is related to the matched gene entry. Each approved symbol is hyperlinked to the HGNC Symbol Report so that users can view further information on the matched genes.
Similar articles
- Genenames.org: the HGNC resources in 2023.
Seal RL, Braschi B, Gray K, Jones TEM, Tweedie S, Haim-Vilmovsky L, Bruford EA. Seal RL, et al. Nucleic Acids Res. 2023 Jan 6;51(D1):D1003-D1009. doi: 10.1093/nar/gkac888. Nucleic Acids Res. 2023. PMID: 36243972 Free PMC article. - Gene family matters: expanding the HGNC resource.
Daugherty LC, Seal RL, Wright MW, Bruford EA. Daugherty LC, et al. Hum Genomics. 2012 Jul 5;6(1):4. doi: 10.1186/1479-7364-6-4. Hum Genomics. 2012. PMID: 23245209 Free PMC article. Review. - Genenames.org: the HGNC and VGNC resources in 2017.
Yates B, Braschi B, Gray KA, Seal RL, Tweedie S, Bruford EA. Yates B, et al. Nucleic Acids Res. 2017 Jan 4;45(D1):D619-D625. doi: 10.1093/nar/gkw1033. Epub 2016 Oct 30. Nucleic Acids Res. 2017. PMID: 27799471 Free PMC article. - The HGNC Database in 2008: a resource for the human genome.
Bruford EA, Lush MJ, Wright MW, Sneddon TP, Povey S, Birney E. Bruford EA, et al. Nucleic Acids Res. 2008 Jan;36(Database issue):D445-8. doi: 10.1093/nar/gkm881. Epub 2007 Nov 4. Nucleic Acids Res. 2008. PMID: 17984084 Free PMC article. - A review of the new HGNC gene family resource.
Gray KA, Seal RL, Tweedie S, Wright MW, Bruford EA. Gray KA, et al. Hum Genomics. 2016 Feb 3;10:6. doi: 10.1186/s40246-016-0062-6. Hum Genomics. 2016. PMID: 26842383 Free PMC article. Review.
Cited by
- Integrating pathways of Parkinson's disease in a molecular interaction map.
Fujita KA, Ostaszewski M, Matsuoka Y, Ghosh S, Glaab E, Trefois C, Crespo I, Perumal TM, Jurkowski W, Antony PM, Diederich N, Buttini M, Kodama A, Satagopam VP, Eifes S, Del Sol A, Schneider R, Kitano H, Balling R. Fujita KA, et al. Mol Neurobiol. 2014 Feb;49(1):88-102. doi: 10.1007/s12035-013-8489-4. Epub 2013 Jul 7. Mol Neurobiol. 2014. PMID: 23832570 Free PMC article. Review. - HIPPIE: Integrating protein interaction networks with experiment based quality scores.
Schaefer MH, Fontaine JF, Vinayagam A, Porras P, Wanker EE, Andrade-Navarro MA. Schaefer MH, et al. PLoS One. 2012;7(2):e31826. doi: 10.1371/journal.pone.0031826. Epub 2012 Feb 14. PLoS One. 2012. PMID: 22348130 Free PMC article. - Dr.VIS: a database of human disease-related viral integration sites.
Zhao X, Liu Q, Cai Q, Li Y, Xu C, Li Y, Li Z, Zhang X. Zhao X, et al. Nucleic Acids Res. 2012 Jan;40(Database issue):D1041-6. doi: 10.1093/nar/gkr1142. Epub 2011 Dec 1. Nucleic Acids Res. 2012. PMID: 22135288 Free PMC article. - Proline: the distribution, frequency, positioning, and common functional roles of proline and polyproline sequences in the human proteome.
Morgan AA, Rubenstein E. Morgan AA, et al. PLoS One. 2013;8(1):e53785. doi: 10.1371/journal.pone.0053785. Epub 2013 Jan 25. PLoS One. 2013. PMID: 23372670 Free PMC article. - Community gene annotation in practice.
Loveland JE, Gilbert JG, Griffiths E, Harrow JL. Loveland JE, et al. Database (Oxford). 2012 Mar 20;2012:bas009. doi: 10.1093/database/bas009. Print 2012. Database (Oxford). 2012. PMID: 22434843 Free PMC article.
References
- Shows TB, Alper CA, Bootsma D, Dorf M, Douglas T, Huisman T, Kit S, Klinger HP, Kozak C, Lalley PA, et al. International system for human gene nomenclature (1979) ISGN (1979) Cytogenet. Cell Genet. 1979;25:96–116. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical