Improved variant discovery through local re-alignment of short-read next-generation sequencing data using SRMA - PubMed (original) (raw)
Improved variant discovery through local re-alignment of short-read next-generation sequencing data using SRMA
Nils Homer et al. Genome Biol. 2010.
Abstract
A primary component of next-generation sequencing analysis is to align short reads to a reference genome, with each read aligned independently. However, reads that observe the same non-reference DNA sequence are highly correlated and can be used to better model the true variation in the target genome. A novel short-read micro realigner, SRMA, that leverages this correlation to better resolve a consensus of the underlying DNA sequence of the targeted genome is described here.
Figures
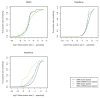
Figure 1
Local re-alignment receiver operator characteristic curves for simulated human genome re-sequencing data. A synthetic diploid human genome with SNPs, deletions, and insertions was created from a reference human genome (hg18) as described in main text. One billion paired 50-mer reads for both base space and color space were simulated from this synthetic genome to assess the true positive and false positive rates of variant calling after re-sequencing. An increasing SNP quality filter was used to generate each curve. The simulated dataset was aligned with BWA (v.0.5.7-5) with the default parameters [9]. The alignments from BWA and SRMA were variant called using the MAQ consensus model implemented in SAMtools (v.0.1.17) using the default settings [10,20]. For the simulated datasets, the resulting variant calls were assessed for accuracy by comparing the called variants against the known introduced sites of variation. The BWA alignments were locally re-aligned with SRMA with variant inclusive settings (c = 2 and p = 0.1).
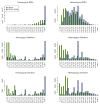
Figure 2
Allele frequency distribution with local re-alignment of U87MG. SRMA was applied to the alignments produced with BFAST of a human cancer cell line (U87MG; SRA009912.1). Variants were called with SAMtools before and after application of SRMA (see Materials and methods). Homozygous and heterozygous calls were examined independently using zygosity calls produced by SAMtools. The observed non-reference allele frequency for SNPs, deletions, and insertions are plotted for homozygous (left panels) or heterozygous variants (right panels). Ideally, non-reference allele frequencies for homozygous and heterozygous variants approach 1.0 and 0.5, respectively. The absolute counts of observed variants are plotted (y-axis) against non-reference allele frequency ranges (x-axis).
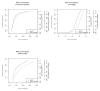
Figure 3
dbSNP concordance before and after local re-alignment of U87MG. SRMA was applied to the alignments produced with BFAST of a human cancer cell line (U87MG; SRA009912.1). Variants were called with SAMtools before and after application of SRMA (see Materials and methods). Deletions and insertions (indels) called within U87MG were compared with those indels reported in dbSNP (v129). An increasing minimum SNP quality filter was used to improve concordance (y-axis) while reducing the number of indels observed at dbSNP positions (x-axis). Using SRMA significantly reduced the discordance (one minus concordance) between observed indels at dbSNP positions.
Figure 4
SNP microarray concordance with known genotypes before and after local re-alignment of U87MG. SRMA was applied to the alignments produced with BFAST of a human cancer cell line (U87MG; SRA009912.1). Heterozygous genotypes from an Illumina SNP microarray were compared with genotypes called from sequence data before and after application of SRMA (see Materials and methods). A minimum threshold on three different variant-calling metrics was applied, respectively, to improve the concordance (y-axis) while reducing the total number of SNP positions on the microarray that were called. Regardless of the metric, SRMA reduced the discordance (one minus concordance) of heterozygous SNPs reported by the SNP microarray and sequencing data.
Figure 5
A deletion and SNP in ALPK2 in U87MG. SRMA was applied to the alignments produced with BFAST of a human cancer cell line (U87MG; SRA009912.1). (a,b) The resulting alignments from within the coding region of ALPK2 (chr18:54,355,303-54,355,477) are shown before applying SRMA (a) and after applying SRMA (b). In this haploid region, Sanger sequencing confirmed a 15-bp deletion and a C-to-T SNP eight bases downstream of the deletion. Panel (a) shows the difficulty of aligning sequence reads from a region with a large deletion and a SNP, as false variation is observed (SNPs and indels). Nevertheless, some reads in (a) (BFAST) do correctly observe the deletion and SNP, which are therefore included in the variant graph created by SRMA. After local re-alignment using SRMA (b), the majority of the reads support the presence of the deletion and SNP, while false variation has been eliminated. The Integrated Genomics Viewer was used to view the alignments [30].
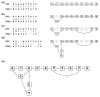
Figure 6
The creation of a variant graph. Four alignments (left) are successively used to create a variant graph (right). (a) An alignment of a read that matches the reference. The associated variant graph consists of nodes that represent each base of the read. (b) An alignment of a read with a base difference at the second position. The base difference adds a new node that is connected to the existing first and third node. (c) An alignment of a read that has a base difference and a deletion relative to the reference. A new edge connecting the sixth and ninth nodes is added to the graph. (d) An alignment of a read that has a base difference, a deletion, and an insertion relative to the reference. Two new nodes are added creating a path from the previously existing SNP at the second position to the reference base at the second position. (e) The resulting variant graph with each edge labeled with the number of alignment paths containing this edge.
Similar articles
- ABRA: improved coding indel detection via assembly-based realignment.
Mose LE, Wilkerson MD, Hayes DN, Perou CM, Parker JS. Mose LE, et al. Bioinformatics. 2014 Oct;30(19):2813-5. doi: 10.1093/bioinformatics/btu376. Epub 2014 Jun 6. Bioinformatics. 2014. PMID: 24907369 Free PMC article. - STR-realigner: a realignment method for short tandem repeat regions.
Kojima K, Kawai Y, Misawa K, Mimori T, Nagasaki M. Kojima K, et al. BMC Genomics. 2016 Dec 3;17(1):991. doi: 10.1186/s12864-016-3294-x. BMC Genomics. 2016. PMID: 27912743 Free PMC article. - Analysis of high-throughput sequencing data.
Mane SP, Modise T, Sobral BW. Mane SP, et al. Methods Mol Biol. 2011;678:1-11. doi: 10.1007/978-1-60761-682-5_1. Methods Mol Biol. 2011. PMID: 20931368 - A survey of sequence alignment algorithms for next-generation sequencing.
Li H, Homer N. Li H, et al. Brief Bioinform. 2010 Sep;11(5):473-83. doi: 10.1093/bib/bbq015. Epub 2010 May 11. Brief Bioinform. 2010. PMID: 20460430 Free PMC article. Review. - Computational methods for discovering structural variation with next-generation sequencing.
Medvedev P, Stanciu M, Brudno M. Medvedev P, et al. Nat Methods. 2009 Nov;6(11 Suppl):S13-20. doi: 10.1038/nmeth.1374. Nat Methods. 2009. PMID: 19844226 Review.
Cited by
- Risk and Resilience Variants in the Retinoic Acid Metabolic and Developmental Pathways Associated with Risk of FASD Outcomes.
McKay L, Petrelli B, Pind M, Reynolds JN, Wintle RF, Chudley AE, Drögemöller B, Fainsod A, Scherer SW, Hanlon-Dearman A, Hicks GG. McKay L, et al. Biomolecules. 2024 May 10;14(5):569. doi: 10.3390/biom14050569. Biomolecules. 2024. PMID: 38785976 Free PMC article. - Genotyping of the rare Para-Bombay blood group in southern Thailand.
Rattanapan Y, Charong N, Narkpetch S, Chareonsirisuthigul T. Rattanapan Y, et al. Hematol Transfus Cell Ther. 2023 Oct-Dec;45(4):449-455. doi: 10.1016/j.htct.2022.08.004. Epub 2022 Oct 9. Hematol Transfus Cell Ther. 2023. PMID: 36241527 Free PMC article. - Higher genome mutation rates of Beijing lineage of Mycobacterium tuberculosis during human infection.
Hakamata M, Takihara H, Iwamoto T, Tamaru A, Hashimoto A, Tanaka T, Kaboso SA, Gebretsadik G, Ilinov A, Yokoyama A, Ozeki Y, Nishiyama A, Tateishi Y, Moro H, Kikuchi T, Okuda S, Matsumoto S. Hakamata M, et al. Sci Rep. 2020 Oct 22;10(1):17997. doi: 10.1038/s41598-020-75028-2. Sci Rep. 2020. PMID: 33093577 Free PMC article. - Comprehensive assay for the molecular profiling of cancer by target enrichment from formalin-fixed paraffin-embedded specimens.
Kohsaka S, Tatsuno K, Ueno T, Nagano M, Shinozaki-Ushiku A, Ushiku T, Takai D, Ikegami M, Kobayashi H, Kage H, Ando M, Hata K, Ueda H, Yamamoto S, Kojima S, Oseto K, Akaike K, Suehara Y, Hayashi T, Saito T, Takahashi F, Takahashi K, Takamochi K, Suzuki K, Nagayama S, Oda Y, Mimori K, Ishihara S, Yatomi Y, Nagase T, Nakajima J, Tanaka S, Fukayama M, Oda K, Nangaku M, Miyazono K, Miyagawa K, Aburatani H, Mano H. Kohsaka S, et al. Cancer Sci. 2019 Apr;110(4):1464-1479. doi: 10.1111/cas.13968. Epub 2019 Mar 5. Cancer Sci. 2019. PMID: 30737998 Free PMC article. - Jointly aligning a group of DNA reads improves accuracy of identifying large deletions.
Shrestha AMS, Frith MC, Asai K, Richard H. Shrestha AMS, et al. Nucleic Acids Res. 2018 Feb 16;46(3):e18. doi: 10.1093/nar/gkx1175. Nucleic Acids Res. 2018. PMID: 29182778 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
- R01 MH071852/MH/NIMH NIH HHS/United States
- U24 NS052108/NS/NINDS NIH HHS/United States
- U01HG005210/HG/NHGRI NIH HHS/United States
- U24NS052108/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources