The piRNA pathway: a fly's perspective on the guardian of the genome - PubMed (original) (raw)
Review
The piRNA pathway: a fly's perspective on the guardian of the genome
Kirsten-André Senti et al. Trends Genet. 2010 Dec.
Abstract
Throughout the eukaryotic lineage, small RNA silencing pathways protect the genome against the deleterious influence of selfish genetic elements such as transposons. In animals an elaborate small RNA pathway centered on PIWI proteins and their interacting piRNAs silences transposons within the germline. In contrast to other small RNA silencing pathways, we lack a mechanistic understanding of this genome defense system. However, genetic and molecular studies have uncovered a fascinating conceptual framework for this pathway that is conserved from sponges to mammals. We discuss our current understanding of the piRNA pathway in Drosophila with an emphasis on origin and biogenesis of piRNAs.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures
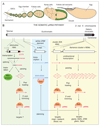
Figure 1. A primary piRNA pathway is active in somatic cells of the Drosophila ovary
A) The Drosophila oocyte is in direct contact with germline-derived cells (beige) and is surrounded by cells of somatic origin (green). This cartoon depicts an ovariole, the functional unit of the ovary (reproduced with kind permission from A. Spradling 110). Development proceeds from left (germarium) to right (mature egg). In the germarium, Germline Stem Cells (GSCs) divide asymmetrically into GSCs and differentiating cystoblasts. Four mitotic cystoblast cell divisions produce 15 nurse cells and an oocyte that remain connected by cytoplasmic bridges. Each of these germline cell clusters is surrounded by an epithelium of somatic follicle cells (green) to form an egg chamber that continuously grows until the oocyte matures into an egg. Follicle cells finally undergo apoptosis after depositing the eggshell. The deposited egg therefore lacks somatic cells. B) Shown is a schematic representation of the somatic piRNA pathway (primary piRNA module). For illustrative purposes, piRNA source and target loci from the X-chromosome (drawn at the top) are displayed. Colored boxes summarize primary piRNA biogenesis from piRNA clusters (yellow) and from 3’UTRs of protein coding genes (green). The blue box shows expression and silencing of ZAM, a prototypical LTR-retrotransposon, active in follicle cells. (yellow box) The piRNA clusters 20A and flamenco are located at the boundary between euchromatin and heterochromatin. Both contain almost exclusively transposon fragments oriented antisense to the unidirectional promoter. piRNA cluster transcripts (red) therefore give rise to antisense piRNAs. Unknown mechanisms parse piRNA cluster transcripts into shorter fragments that might enter Piwi. At this step, Piwi could preferentially select precursors with a 5’ Uridine (1U). Subsequently, the 3’ tail of Piwi bound RNAs is trimmed and 2’OH-methylated to generate mature piRNAs. (blue box) The sequence of mature piRNAs defines their target: Displayed is an active copy of the ZAM LTR-retrotransposon and its sense transcript (blue box). The green box summarizes piRNA biogenesis from genes (here diminuitive). The spliced dm transcript with 5’UTR, coding sequence and 3’UTR is shown. Mature dm mRNAs are either translated into Myc or act as piRNA precursors. piRNAs are preferentially processed from 3’UTR sequences, presumably by a similar mechanism as for piRNA cluster transcripts. Genic piRNAs are in sense orientation to the host gene and their targets (if any) remain to be identified.
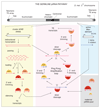
Figure 2. In germline cells, the primary piRNA pathway and the Ping-Pong amplification loop are active
Shown are representative examples of germline piRNA sources and targets originating from the 2nd chromosome (drawn at the top). Colored boxes show primary piRNA biogenesis from cluster 42AB (yellow), the adaptive module of the target dependent ping-pong amplification loop (red), expression and silencing of a typical active LTR-retrotransposon (_Max-_Element; blue) and the contribution of maternally inherited piRNAs (green). (yellow box) Cluster 42AB contains transposon fragments in both orientations and is bi-directionally transcribed. During primary piRNA biogenesis, cluster transcripts (red) presumably generate sense and antisense piRNAs. Unknown mechanisms parse the long piRNA precursor transcripts into shorter fragments that are loaded onto PIWI family proteins (Piwi, probably Aub and potentially AGO3). Piwi and Aub probably select RNA fragments with a 5’ Uridine (1U). Subsequently, the 3’ tail of pre-piRNAs are trimmed and 2’OH-methylated to generate mature piRNAs. piRISCs with antisense piRNAs are competent to silence sequence complementary transcripts of active transposons. Primary piRNA biogenesis in the germline is likely similar to the one in somatic cells. The blue box shows an active copy of the _Max-_Element (LTR-retrotransposon) and its transcribed sense transcript that is silenced by complementary piRISCs. The red box summarizes the ping-pong cycle. An Aub complexed piRNA (red) that is antisense to an active sense Max transcript (dark blue) guides slicing (scissors) of the transposon RNA, precisely 10nt downstream of its 5’ Uridine. The sliced Max transcript is predicted to be loaded onto AGO3 and typically has a profound bias for an Adenine at position 10 (10A). The AGO3 bound pre-piRNA is 3’ trimmed and 2’-OH methylated. This mature AGO3-piRNA complex in turn cleaves complementary cluster transcripts and triggers production of an Aub-loaded antisense piRNA, whose sequence is identical to the initiating piRNA. It is currently impossible to experimentally distinguish between Aub-piRNA complexes generated via primary piRNA biogenesis or via ping-pong. Weak ping-pong signatures exist between AGO3 and Piwi and could indicate that Piwi (besides primary biogenesis) also receives piRNAs via AGO3 mediated target slicing. (green box) At the end of oogenesis, mature Aub- and Piwi-piRNA complexes (to a lesser extent also AGO3) are efficiently loaded into the oocyte. Maternal Aub and to a lower extent also Piwi localize to the posterior pole of the mature egg, where the future germline will form. Maternal piRNAs might serve important roles in the starting phase of the pig-pong cycle.
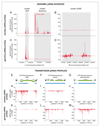
Figure 3. piRNA profiles along clusters and transposons are tissue specific
This figure illustrates the pronounced differences in piRNA pools found in somatic and germline cells of the Drosophila ovary. Basis for these differences are tissue specific transcription of piRNA clusters and presence of the ping-pong cycle in germline cells only. Somatic graphs are based on the OSS cell data from ref.43 and germline graphs are based on early embryo libraries from ref.66. To enable comparison of these populations, profiles were always normalized to 1 millionn sequenced repeat-derived 23-30nt small RNAs. Panels A-D indicate that flamenco is a soma-specific piRNA cluster while cluster 42AB is germline specific (cluster coordinates are shaded in light grey). Cluster 20A is processed into piRNAs in both cell-types. Also apparent is the unidirectional transcription of flamenco and cluster 20A while cluster 42AB is transcribed in both orientations. In each panel, only genome-unique piRNAs were used and a 200 nt sliding window with step size of 20 nt was applied. Sense and antisense piRNAs are displayed as upwards and downwards peaks, respectively (E-G) Shown are schematics of the LTR retrotransposon ZAM, Blood and _Max_-Element. Blue bars display the respective transposon fragments found in piRNA clusters (antisense ZAM fragments within flamenco, a complete antisense Blood element in cluster20A and Max fragments in cluster 42AB). Transposon cartoons and cluster fragments are length matched to the piRNA profiles shown below. Panels H-M show profiles of somatic and germline piRNAs mapping to ZAM, Blood and Max. For each graph, piRNAs mapping with up to three mismatches to the indicated element were pooled. ZAM is a proto-typical element expressed and silenced in somatic cells, while Max is apparently only silenced (and presumably transcribed) in germline cells. Blood silencing is active in both cell-types. The ZAM fragments present in the flamenco piRNA cluster (blue) are in striking agreement with the observed piRNA profiles, suggesting that they are the major source of ZAM piRNAs. Similarly, piRNA profiles for Blood and Max are consistent with their respective fragments in piRNA clusters 20A and 42AB. Ping-pong signatures are significant only for Blood and Max in the germline samples (not shown).
Similar articles
- The piRNA pathway in Drosophila ovarian germ and somatic cells.
Sato K, Siomi MC. Sato K, et al. Proc Jpn Acad Ser B Phys Biol Sci. 2020;96(1):32-42. doi: 10.2183/pjab.96.003. Proc Jpn Acad Ser B Phys Biol Sci. 2020. PMID: 31932527 Free PMC article. Review. - Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary.
Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Malone CD, et al. Cell. 2009 May 1;137(3):522-35. doi: 10.1016/j.cell.2009.03.040. Epub 2009 Apr 23. Cell. 2009. PMID: 19395010 Free PMC article. - Transposon silencing by piRNAs.
Halic M, Moazed D. Halic M, et al. Cell. 2009 Sep 18;138(6):1058-60. doi: 10.1016/j.cell.2009.08.030. Cell. 2009. PMID: 19766558 Free PMC article. - piRNA-mediated silencing in Drosophila germlines.
Siomi MC, Miyoshi T, Siomi H. Siomi MC, et al. Semin Cell Dev Biol. 2010 Sep;21(7):754-9. doi: 10.1016/j.semcdb.2010.01.011. Epub 2010 Jan 18. Semin Cell Dev Biol. 2010. PMID: 20080197 Review. - The genetic makeup of the Drosophila piRNA pathway.
Handler D, Meixner K, Pizka M, Lauss K, Schmied C, Gruber FS, Brennecke J. Handler D, et al. Mol Cell. 2013 Jun 6;50(5):762-77. doi: 10.1016/j.molcel.2013.04.031. Epub 2013 May 9. Mol Cell. 2013. PMID: 23665231 Free PMC article.
Cited by
- Function of Piwi, a nuclear Piwi/Argonaute protein, is independent of its slicer activity.
Darricarrère N, Liu N, Watanabe T, Lin H. Darricarrère N, et al. Proc Natl Acad Sci U S A. 2013 Jan 22;110(4):1297-302. doi: 10.1073/pnas.1213283110. Epub 2013 Jan 7. Proc Natl Acad Sci U S A. 2013. PMID: 23297219 Free PMC article. - Regulatory RNAs in the light of Drosophila genomics.
Marco A. Marco A. Brief Funct Genomics. 2012 Sep;11(5):356-65. doi: 10.1093/bfgp/els033. Epub 2012 Sep 5. Brief Funct Genomics. 2012. PMID: 22956639 Free PMC article. Review. - RNA-mediated epigenetic regulation of gene expression.
Holoch D, Moazed D. Holoch D, et al. Nat Rev Genet. 2015 Feb;16(2):71-84. doi: 10.1038/nrg3863. Epub 2015 Jan 2. Nat Rev Genet. 2015. PMID: 25554358 Free PMC article. Review. - Reexamining the P-Element Invasion of Drosophila melanogaster Through the Lens of piRNA Silencing.
Kelleher ES. Kelleher ES. Genetics. 2016 Aug;203(4):1513-31. doi: 10.1534/genetics.115.184119. Genetics. 2016. PMID: 27516614 Free PMC article. Review. - Serum non-coding RNAs as biomarkers for osteoarthritis progression after ACL injury.
Zhang L, Yang M, Marks P, White LM, Hurtig M, Mi QS, Divine G, Gibson G. Zhang L, et al. Osteoarthritis Cartilage. 2012 Dec;20(12):1631-7. doi: 10.1016/j.joca.2012.08.016. Epub 2012 Aug 31. Osteoarthritis Cartilage. 2012. PMID: 22944527 Free PMC article.
References
- Hurst GD, Werren JH. The role of selfish genetic elements in eukaryotic evolution. Nat Rev Genet. 2001;2:597–606. - PubMed
- Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. - PubMed
- Bucheton A, et al. The molecular basis of I-R hybrid dysgenesis in Drosophila melanogaster: identification, cloning, and properties of the I factor. Cell. 1984;38:153–163. - PubMed
- Rubin GM, et al. The molecular basis of P-M hybrid dysgenesis: the nature of induced mutations. Cell. 1982;29:987–994. - PubMed
- Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases