Countering amyloid polymorphism and drug resistance with minimal drug cocktails - PubMed (original) (raw)
Countering amyloid polymorphism and drug resistance with minimal drug cocktails
Martin L Duennwald et al. Prion. 2010 Oct-Dec.
Abstract
Several fatal, progressive neurodegenerative diseases, including various prion and prion-like disorders, are connected with the misfolding of specific proteins. These proteins misfold into toxic oligomeric species and a spectrum of distinct self-templating amyloid structures, termed strains. Hence, small molecules that prevent or reverse these protein-misfolding events might have therapeutic utility. Yet it is unclear whether a single small molecule can antagonize the complete repertoire of misfolded forms encompassing diverse amyloid polymorphs and soluble oligomers. We have begun to investigate this issue using the yeast prion protein Sup35 as an experimental paradigm. We have discovered that a polyphenol, (-)epigallocatechin-3-gallate (EGCG), effectively inhibited the formation of infectious amyloid forms (prions) of Sup35 and even remodeled preassembled prions. Surprisingly, EGCG selectively modulated specific prion strains and even selected for EGCG-resistant prion strains with novel structural and biological characteristics. Thus, treatment with a single small molecule antagonist of amyloidogenesis can select for novel, drug-resistant amyloid polymorphs. Importantly, combining EGCG with another small molecule, 4,5-bis-(4-methoxyanilino)phthalimide, synergistically antagonized and remodeled a wide array of Sup35 prion strains without producing any drug-resistant prions. We suggest that minimal drug cocktails, small collections of drugs that collectively antagonize all amyloid polymorphs, should be identified to besiege various neurodegenerative disorders.
Figures
Figure 1
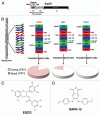
Sup35 prion strains and small-molecule antagonists. (A) Sup35 is a modular protein comprised of a C-terminal GTPase domain (C, amino acids 254–685, black), a highly charged middle domain (M, amino acids 124–253, dark grey) and an N-terminal domain (N, amino acids 1–123, light grey) enriched in glutamine, asparagine, tyrosine and glycine residues. Together N and M (NM) confer all the properties needed to form a stable prion in yeast. NM is termed the prion domain. Within N, prion recognition elements termed the “head” (red) and “tail” (green), which flank a “central core” (blue), play important roles in prion formation., (B) Sup35 prions adopt a polymeric cross-beta structure. In one proposed model (left), this amyloid structure is composed of the head (red), central core (blue) and tail (green) regions of N. The M and C domains are located on the exterior of this structure and are not depicted for clarity. If we zoom in on three adjacent monomers in the Sup35 prion polymer, we find that the prion is proposed to be maintained by an alternating sequence of head-to-head (red) and tail-to-tail (green) intermolecular contacts. The central core is sequestered in intramolecular contacts (blue). Different Sup35 prion strains assemble under different environmental conditions. Thus, NM25 assembles at 25°C or when NM is chemically crosslinked with BMB in the tail region., NM4 assembles at 4°C or when NM is chemically crosslinked with BMB in the head region., NM4E assembles in the presence of EGCG at 4°C. These prion strains have subtle differences in the precise residues that comprise the head, tail and central core (right). The residues that comprise the head, tail and central core are shown to the right of each central protomer. NM25, NM4 and NM4E are distinguished by their different tail-to-tail contacts and central core region. Moreover, NM4E has a distinct head-to-head contact. Infection of [_psi_−] [_pin_−] cells with NM25 yields mostly weak [PSI+] variants, whereas NM4 yields mostly strong [PSI+] variants. By contrast, NM4E generates purely strong [PSI+] variants (pie charts in lower portion). (C) Chemical structure of EGCG. (D) Chemical structure of DAPH-12.
Similar articles
- A synergistic small-molecule combination directly eradicates diverse prion strain structures.
Roberts BE, Duennwald ML, Wang H, Chung C, Lopreiato NP, Sweeny EA, Knight MN, Shorter J. Roberts BE, et al. Nat Chem Biol. 2009 Dec;5(12):936-46. doi: 10.1038/nchembio.246. Epub 2009 Nov 1. Nat Chem Biol. 2009. PMID: 19915541 Free PMC article. - Radically different amyloid conformations dictate the seeding specificity of a chimeric Sup35 prion.
Foo CK, Ohhashi Y, Kelly MJ, Tanaka M, Weissman JS. Foo CK, et al. J Mol Biol. 2011 Apr 22;408(1):1-8. doi: 10.1016/j.jmb.2011.02.025. Epub 2011 Feb 17. J Mol Biol. 2011. PMID: 21333653 Free PMC article. - Direct and selective elimination of specific prions and amyloids by 4,5-dianilinophthalimide and analogs.
Wang H, Duennwald ML, Roberts BE, Rozeboom LM, Zhang YL, Steele AD, Krishnan R, Su LJ, Griffin D, Mukhopadhyay S, Hennessy EJ, Weigele P, Blanchard BJ, King J, Deniz AA, Buchwald SL, Ingram VM, Lindquist S, Shorter J. Wang H, et al. Proc Natl Acad Sci U S A. 2008 May 20;105(20):7159-64. doi: 10.1073/pnas.0801934105. Epub 2008 May 14. Proc Natl Acad Sci U S A. 2008. PMID: 18480256 Free PMC article. - [Mechanism and application of molecular self-assembly in Sup35 prion domain of Saccharomyces cerevisiae].
Yin W, He J, Yu Z, Wang J. Yin W, et al. Sheng Wu Gong Cheng Xue Bao. 2011 Oct;27(10):1401-7. Sheng Wu Gong Cheng Xue Bao. 2011. PMID: 22260056 Review. Chinese. - Emergence and natural selection of drug-resistant prions.
Shorter J. Shorter J. Mol Biosyst. 2010 Jul;6(7):1115-30. doi: 10.1039/c004550k. Epub 2010 Apr 27. Mol Biosyst. 2010. PMID: 20422111 Free PMC article. Review.
Cited by
- The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease.
King OD, Gitler AD, Shorter J. King OD, et al. Brain Res. 2012 Jun 26;1462:61-80. doi: 10.1016/j.brainres.2012.01.016. Epub 2012 Jan 21. Brain Res. 2012. PMID: 22445064 Free PMC article. Review. - The role of amyloidogenic protein oligomerization in neurodegenerative disease.
Lotz GP, Legleiter J. Lotz GP, et al. J Mol Med (Berl). 2013 Jun;91(6):653-64. doi: 10.1007/s00109-013-1025-1. Epub 2013 Mar 27. J Mol Med (Berl). 2013. PMID: 23529761 Review. - Combating deleterious phase transitions in neurodegenerative disease.
Darling AL, Shorter J. Darling AL, et al. Biochim Biophys Acta Mol Cell Res. 2021 Apr;1868(5):118984. doi: 10.1016/j.bbamcr.2021.118984. Epub 2021 Feb 5. Biochim Biophys Acta Mol Cell Res. 2021. PMID: 33549703 Free PMC article. Review. - Hsp104 drives "protein-only" positive selection of Sup35 prion strains encoding strong [PSI(+)].
DeSantis ME, Shorter J. DeSantis ME, et al. Chem Biol. 2012 Nov 21;19(11):1400-10. doi: 10.1016/j.chembiol.2012.09.013. Chem Biol. 2012. PMID: 23177195 Free PMC article. - The Surprising Role of Amyloid Fibrils in HIV Infection.
Castellano LM, Shorter J. Castellano LM, et al. Biology (Basel). 2012 May 29;1(1):58-80. doi: 10.3390/biology1010058. Biology (Basel). 2012. PMID: 24832047 Free PMC article.
References
- Soto C, Estrada LD. Protein misfolding and neurodegeneration. Arch Neurol. 2008;65:184–189. - PubMed
- Forman MS, Trojanowski JQ, Lee VM. Neurodegenerative diseases: a decade of discoveries paves the way for therapeutic breakthroughs. Nat Med. 2004;10:1055–1063. - PubMed
- Nelson R, Eisenberg D. Structural models of amyloid-like fibrils. Adv Protein Chem. 2006;73:235–282. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous