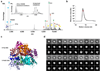The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations - PubMed (original) (raw)
. 2010 Nov;17(11):1324-9.
doi: 10.1038/nsmb.1920. Epub 2010 Oct 10.
Jin Kuk Yang, Venkataraman Kabaleeswaran, Amanda J Rice, Anthony C Cruz, Ah Young Park, Qian Yin, Ermelinda Damko, Se Bok Jang, Stefan Raunser, Carol V Robinson, Richard M Siegel, Thomas Walz, Hao Wu
Affiliations
- PMID: 20935634
- PMCID: PMC2988912
- DOI: 10.1038/nsmb.1920
The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations
Liwei Wang et al. Nat Struct Mol Biol. 2010 Nov.
Abstract
The death-inducing signaling complex (DISC) formed by the death receptor Fas, the adaptor protein FADD and caspase-8 mediates the extrinsic apoptotic program. Mutations in Fas that disrupt the DISC cause autoimmune lymphoproliferative syndrome (ALPS). Here we show that the Fas-FADD death domain (DD) complex forms an asymmetric oligomeric structure composed of 5-7 Fas DD and 5 FADD DD, whose interfaces harbor ALPS-associated mutations. Structure-based mutations disrupt the Fas-FADD interaction in vitro and in living cells; the severity of a mutation correlates with the number of occurrences of a particular interaction in the structure. The highly oligomeric structure explains the requirement for hexameric or membrane-bound FasL in Fas signaling. It also predicts strong dominant negative effects from Fas mutations, which are confirmed by signaling assays. The structure optimally positions the FADD death effector domain (DED) to interact with the caspase-8 DED for caspase recruitment and higher-order aggregation.
Figures
Fig. 1
Biochemical and structural characterization of the Fas DD–FADD DD complexes. (a) Tandem mass spectrum of the hFas–hFADD complex showing the dissociated monomers and the ‘stripped’ complexes at high gas phase collisional activation. The peak centered at 5913 m/z was isolated and collision energy (50–100 V) was applied. Well-resolved peaks at high m/z correspond to the stripped complexes for the 5:5, 6:5 and 7:5 complexes. Inset, mass spectrum of the hFas–hFADD complex showing monomers and dimers of hFas and hFADD at lower m/z and the unresolved charge series at higher m/z. (b) A representative multi-angle light scattering (MALS) measurement of the hFas–hFADD complex. More MALS data are summarized in Supplementary Table 1. (c) Crystal structure of the core 5:5 mFas–hFADD complex. The Fas molecules are shown in warm colors and the FADD DD molecules are shown in cold colors. (d) Class averages of the mFas–hFadd complex (1st and 4th lines) with best matching projections from the current mFas–hFADD crystal structure (2nd and 5th lines) and from a previously published structure of the Fas–FADD complex (3rd and 6th lines) .
Fig. 2
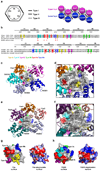
Interactions in the Fas DD–FADD DD complex. (a) Schematic planar diagram showing the construction of the complex. The locations of the three types of contacts are shown. (b) Sequence alignment of human and mouse Fas DD and FADD DD. Major interfacial residues are colored in yellow for type Ia, cyan for type Ib, magenta for type IIa, green for type IIb, red for type IIIa and blue for type IIIb. Single letter codes below the sequences indicate mutations tested with red indicating defective and black indicating non-defective. Locations of ALPS mutations are also shown. (c) Arrangement of molecules around Fas2, viewing into the schematic diagram in (a) from behind the page. (d) Zoom-in of (c) showing residues at the three types of interactions in Fas2 (labeled without a text box) and the surrounding molecules (labeled with a text box). Residues in Fas2 are colored in yellow for type Ia, cyan for type Ib, magenta for type IIa, green for type IIb (residue name not labeled), red for type IIIa and blue for type IIIb. Side chains of residues in the surrounding molecules are shown as stick models colored by atom types. (e) Arrangement of molecules around FADD3, viewing into the schematic diagram in (a) from behind the page. (f) Zoom-in of (e) showing residues at the three types of interactions in FADD3 (labeled without a text box) and the surrounding molecules (labeled with a text box). Residues in FADD3 are colored in yellow for type Ia, cyan for type Ib, magenta for type IIa (residue name not labeled), green for type IIb, red for type IIIa and blue for type IIIb. Side chains of residues in the surrounding molecules are shown as stick models colored by atom types. (g) Side by side surface representations of Fas DD showing the locations of the three types of interactions (left) and the surface charge features (right). (h) Side by side surface representations of FADD DD showing the locations of the three types of interactions (left) and the surface charge features (right).
Fig. 3
Structure-based mutagenesis and analysis of ALPS mutations. (a) Analysis of structure-based mutations of Fas DD and FADD DD in the in vitro His-tag pulldown assay. (−) indicates a defective interaction. PD: pulldown; I: input. (b) Mapping of ALPS-associated mutation residues onto the surface of Fas DD. (c) Pulldown of ALPS-associated Fas DD mutants by His-tagged FADD DD. (−) indicates a defective interaction. PD: pulldown.
Fig. 4
Interactions in living cells and functional effects of Fas DD and FADD DD mutations. (a) 293T cells were transfected with CFP-tagged FADD DD and YFP-tagged wild-type or mutant full-length Fas and analyzed for FRET by flow cytometry. Bi-exponential plots of the FRET signals are shown for cells transfected with the indicated constructs. A plasmid encoding a truncated Fas protein without a death domain (Fas ΔDD) was used as a negative control. Change in geometric mean fluorescence intensity (ΔMFI) was calculated by subtracting the MFI of non-interacting proteins (Fas ΔDD with WT FADD DD) from the MFI in the FRET channel of the indicated interacting pair. At right the mean fluorescence intensity relative to FRET signal from the Fas ΔDD–FADD interaction (ΔMFI) is shown for the indicated Fas DD mutants interacting with wild-type FADD. The occurrences of the interactions in the complex are shown. (b) FRET analysis of 293T cells transfected with either wild-type or mutant FADD DD interacting with full-length Fas or Fas ΔDD. At right the ΔMFI is shown for the indicated FADD DD mutants interacting with wild-type Fas. The occurrences of the interactions in the complex are shown. Results shown in (a) and (b) are representative of three independent experiments. (c) Effect of Fas DD mutants on Fas-induced cell death in transfected Jurkat E6.1 cells. Cells were transfected with the indicated constructs and apoptosis was induced with anti-Fas or FasL-LZ. Cell death was assayed by flow cytometry. Results are the average +/− s.e.m. of specific cell death in three independent experiments. (d) The Fas DD–full-length FADD complex model constructed by superimposing FADD DD with the structure of full-length FADD . Fas DDs are shown in ribbon diagrams while FADD molecules are shown in surface representations. The location of the FADD DED for caspase-8 recruitment is shown for one of the FADD molecules. (e) A proposed model for post-receptor DISC formation that utilizes FasL hexamerization. All structures are shown in ribbon diagrams except for the intracellular death domains of Fas, which are shown in surface representations. The two FasL trimers are shown in gray. The six Fas extracellular domains are shown in the same color as their intracellular death domains. A 6th Fas intracellular death domain (yellow) was added to the crystal structure. The extracellular and the intracellular domains are connected via straight lines of the same colors. Full-length FADD molecules are shown in ribbon diagrams. The light green bar represents the cellular membrane.
Comment in
- Unleashing cell death: the Fas-FADD complex.
Hymowitz SG, Dixit VM. Hymowitz SG, et al. Nat Struct Mol Biol. 2010 Nov;17(11):1289-90. doi: 10.1038/nsmb1110-1289. Nat Struct Mol Biol. 2010. PMID: 21088666 No abstract available.
Similar articles
- Assembly and activation of the death-inducing signaling complex.
Fosuah E, Shen Z, Xie J, Wang C, Lin Q, Fu TM. Fosuah E, et al. Proc Natl Acad Sci U S A. 2025 Jun 10;122(23):e2504819122. doi: 10.1073/pnas.2504819122. Epub 2025 Jun 4. Proc Natl Acad Sci U S A. 2025. PMID: 40465623 Free PMC article. - Cryo-EM Structure of Caspase-8 Tandem DED Filament Reveals Assembly and Regulation Mechanisms of the Death-Inducing Signaling Complex.
Fu TM, Li Y, Lu A, Li Z, Vajjhala PR, Cruz AC, Srivastava DB, DiMaio F, Penczek PA, Siegel RM, Stacey KJ, Egelman EH, Wu H. Fu TM, et al. Mol Cell. 2016 Oct 20;64(2):236-250. doi: 10.1016/j.molcel.2016.09.009. Epub 2016 Oct 13. Mol Cell. 2016. PMID: 27746017 Free PMC article. - Structural determinants of DISC function: new insights into death receptor-mediated apoptosis signalling.
Sessler T, Healy S, Samali A, Szegezdi E. Sessler T, et al. Pharmacol Ther. 2013 Nov;140(2):186-99. doi: 10.1016/j.pharmthera.2013.06.009. Epub 2013 Jul 8. Pharmacol Ther. 2013. PMID: 23845861 Review. - DED or alive: assembly and regulation of the death effector domain complexes.
Riley JS, Malik A, Holohan C, Longley DB. Riley JS, et al. Cell Death Dis. 2015 Aug 27;6(8):e1866. doi: 10.1038/cddis.2015.213. Cell Death Dis. 2015. PMID: 26313917 Free PMC article. Review.
Cited by
- Type I interferons induce apoptosis by balancing cFLIP and caspase-8 independent of death ligands.
Apelbaum A, Yarden G, Warszawski S, Harari D, Schreiber G. Apelbaum A, et al. Mol Cell Biol. 2013 Feb;33(4):800-14. doi: 10.1128/MCB.01430-12. Epub 2012 Dec 10. Mol Cell Biol. 2013. PMID: 23230268 Free PMC article. - Structural characterization of the self-association of the death domain of p75(NTR.).
Qu Q, Chen J, Wang Y, Gui W, Wang L, Fan Z, Jiang T. Qu Q, et al. PLoS One. 2013;8(3):e57839. doi: 10.1371/journal.pone.0057839. Epub 2013 Mar 5. PLoS One. 2013. PMID: 23472109 Free PMC article. - NleB2 from enteropathogenic Escherichia coli is a novel arginine-glucose transferase effector.
Giogha C, Scott NE, Wong Fok Lung T, Pollock GL, Harper M, Goddard-Borger ED, Pearson JS, Hartland EL. Giogha C, et al. PLoS Pathog. 2021 Jun 16;17(6):e1009658. doi: 10.1371/journal.ppat.1009658. eCollection 2021 Jun. PLoS Pathog. 2021. PMID: 34133469 Free PMC article. - Crystal structure of NALP3 protein pyrin domain (PYD) and its implications in inflammasome assembly.
Bae JY, Park HH. Bae JY, et al. J Biol Chem. 2011 Nov 11;286(45):39528-36. doi: 10.1074/jbc.M111.278812. Epub 2011 Aug 31. J Biol Chem. 2011. PMID: 21880711 Free PMC article. - Proliferative versus apoptotic functions of caspase-8 Hetero or homo: the caspase-8 dimer controls cell fate.
van Raam BJ, Salvesen GS. van Raam BJ, et al. Biochim Biophys Acta. 2012 Jan;1824(1):113-22. doi: 10.1016/j.bbapap.2011.06.005. Epub 2011 Jun 16. Biochim Biophys Acta. 2012. PMID: 21704196 Free PMC article. Review.
References
- Kohl A, Grutter MG. Fire and death: the pyrin domain joins the death-domain superfamily. C R Biol. 2004;327:1077–1086. - PubMed
- Chinnaiyan AM, O'Rourke K, Tewari M, Dixit VM. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell. 1995;81:505–512. - PubMed
- Wajant H. The Fas signaling pathway: more than a paradigm. Science. 2002;296:1635–1636. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI050872/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- RR-15301/RR/NCRR NIH HHS/United States
- R01-AI50872/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous