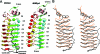Structural and biochemical analysis of the pentapeptide repeat protein EfsQnr, a potent DNA gyrase inhibitor - PubMed (original) (raw)
Structural and biochemical analysis of the pentapeptide repeat protein EfsQnr, a potent DNA gyrase inhibitor
Subray S Hegde et al. Antimicrob Agents Chemother. 2011 Jan.
Abstract
The chromosomally encoded Qnr homolog protein from Enterococcus faecalis (EfsQnr), when expressed, confers to its host a decreased susceptibility to quinolones and consists mainly of tandem repeats, which is consistent with belonging to the pentapeptide repeat family of proteins (PRPs). EfsQnr was cloned with an N-terminal 6× His tag and purified to homogeneity. EfsQnr partially protected DNA gyrase from fluoroquinolone inhibition at concentrations as low as 20 nM. EfsQnr inhibited the ATP-dependent supercoiling activity of DNA gyrase with a 50% inhibitory concentration (IC(50)) of 1.2 μM, while no significant inhibition of ATP-independent relaxation activity was observed. EfsQnr was cytotoxic when overexpressed in Escherichia coli, resulting in the clumping of cells and a loss of viability. The X-ray crystal structure of EfsQnr was determined to 1.6-Å resolution. EfsQnr exhibits the right-handed quadrilateral beta-helical fold typical of PRPs, with features more analogous to MfpA (mycobacterium fluoroquinolone resistance pentapeptide) than to the PRPs commonly found in cyanobacteria.
Figures
FIG. 1.
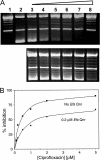
Inhibition of DNA gyrase by _Efs_Qnr. Inhibition of supercoiling (A) and relaxation activities (B) of DNA gyrase by _Efs_Qnr. (A) Lane 1, relaxed plasmid pBR322 alone; lane 2, relaxed pBR322 plus 3 U of gyrase; lanes 3 to 8, 3 U of gyrase and 0.25, 0.5, 1, 2, 5, and 10 μM _Efs_Qnr, respectively. (B) Lane 1, supercoiled plasmid pBR322 alone; lane 2, supercoiled pBR322 plus 50 μM _Efs_Qnr; lane 3, supercoiled pBR322 plus 25 U of gyrase; lanes 4 to 8, 25 U of gyrase and 5, 10, 25, 50, and 100 μM _Efs_Qnr, respectively. nc, l, and sc represent nicked circular, linear, and supercoiled forms, respectively.
FIG. 2.
Inhibition of supercoiling activity by ciprofloxacin and partial protection by _Efs_Qnr. (A) Lane 1, relaxed plasmid pBR322 alone; lane 2, relaxed pBR322 plus 3 U of gyrase; lanes 3 to 8, 3 U of gyrase and 0.1, 0.25, 0.5, 1, 2, and 5 10 μM ciprofloxacin in the absence _Efs_Qnr (top) and in the presence of 0.2 μM _Efs_Qnr (bottom), respectively. (B) Graphical representation of the inhibition data. Symbols are experimentally determined values, while the smooth lines are the fit of the data.
FIG. 3.
Scanning electron microscopy of E. coli BL21(DE3) cells expressing _Efs_Qnr. (A) Control cells; (B) 2 h after induction of _Efs_Qnr expression; (C) 8 h after induction. (D) Effect of _Efs_Qnr induction on the viability of E. coli. Cultures were diluted identically and plated onto LB plates containing 30 μg/ml kanamycin. Bottom, control; top, cells expressing _Efs_Qnr.
FIG. 4.
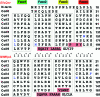
Primary sequence of _Efs_Qnr and _Mt_MfpA. Pentapeptide repeats are compiled into four columns (gray-boxed residues) indicating their location within the four faces of the quadrilateral β-helix. Pentapeptide residue types (i.e., i+1, i+2, etc.) are shown between the two sequences. Helices are displayed in salmon-colored boxes. Residues that support the largest deviations from the standard β-helix are shown in blue.
FIG. 5.
Monomer structure of _Efs_Qnr and _Mt_MfpA. (A) Ribbon diagram of _Efs_Qnr and _Mt_MfpA. β-Helical structural elements are colored by face type as aligned in Fig. 4. α-Helices are in salmon, and the N-terminal extension of _Efs_Qnr is black. Coils have repeats that are colored in the order green, cyan, yellow, and red. Residues within repeats that exhibit full intercoil hydrogen bonding are shown as strands and are indicative of the following turn being type IV, while β-bridges are illustrated as spheres and are indicative of the following turn being type II. (B) Stereoview of the superposition of _Efs_Qnr (black trace) and _Mt_MfpA (orange trace).
FIG. 6.
Molecular dimer of _Efs_Qnr. Ribbon diagram of the _Efs_Qnr and _Mt_MfpA dimer orientated based on the superposition of _Mt_MfpA on subunit B of _Efs_Qnr.
Similar articles
- The pentapeptide repeat proteins MfpAMt and QnrB4 exhibit opposite effects on DNA gyrase catalytic reactions and on the ternary gyrase-DNA-quinolone complex.
Mérens A, Matrat S, Aubry A, Lascols C, Jarlier V, Soussy CJ, Cavallo JD, Cambau E. Mérens A, et al. J Bacteriol. 2009 Mar;191(5):1587-94. doi: 10.1128/JB.01205-08. Epub 2008 Dec 5. J Bacteriol. 2009. PMID: 19060136 Free PMC article. - Structure of QnrB1, a plasmid-mediated fluoroquinolone resistance factor.
Vetting MW, Hegde SS, Wang M, Jacoby GA, Hooper DC, Blanchard JS. Vetting MW, et al. J Biol Chem. 2011 Jul 15;286(28):25265-73. doi: 10.1074/jbc.M111.226936. Epub 2011 May 19. J Biol Chem. 2011. PMID: 21597116 Free PMC article. - Crystal structure of Alr1298, a pentapeptide repeat protein from the cyanobacterium Nostoc sp. PCC 7120, determined at 2.1 Å resolution.
Zhang R, Ni S, Kennedy MA. Zhang R, et al. Proteins. 2020 Sep;88(9):1143-1153. doi: 10.1002/prot.25882. Epub 2020 Feb 24. Proteins. 2020. PMID: 32092202 - Current Understanding of the Structure and Function of Pentapeptide Repeat Proteins.
Zhang R, Kennedy MA. Zhang R, et al. Biomolecules. 2021 Apr 26;11(5):638. doi: 10.3390/biom11050638. Biomolecules. 2021. PMID: 33925937 Free PMC article. Review. - Squaring up to DNA: pentapeptide repeat proteins and DNA mimicry.
Shah S, Heddle JG. Shah S, et al. Appl Microbiol Biotechnol. 2014 Dec;98(23):9545-60. doi: 10.1007/s00253-014-6151-3. Epub 2014 Oct 26. Appl Microbiol Biotechnol. 2014. PMID: 25343976 Review.
Cited by
- Mechanisms of drug resistance: quinolone resistance.
Hooper DC, Jacoby GA. Hooper DC, et al. Ann N Y Acad Sci. 2015 Sep;1354(1):12-31. doi: 10.1111/nyas.12830. Epub 2015 Jul 17. Ann N Y Acad Sci. 2015. PMID: 26190223 Free PMC article. Review. - Structural insights into quinolone antibiotic resistance mediated by pentapeptide repeat proteins: conserved surface loops direct the activity of a Qnr protein from a gram-negative bacterium.
Xiong X, Bromley EH, Oelschlaeger P, Woolfson DN, Spencer J. Xiong X, et al. Nucleic Acids Res. 2011 May;39(9):3917-27. doi: 10.1093/nar/gkq1296. Epub 2011 Jan 11. Nucleic Acids Res. 2011. PMID: 21227918 Free PMC article. - Characterization of Vibrio fluvialis qnrVC5 Gene in Native and Heterologous Hosts: Synergy of qnrVC5 with other Determinants in Conferring Quinolone Resistance.
Vinothkumar K, Kumar GN, Bhardwaj AK. Vinothkumar K, et al. Front Microbiol. 2016 Feb 15;7:146. doi: 10.3389/fmicb.2016.00146. eCollection 2016. Front Microbiol. 2016. PMID: 26913027 Free PMC article. - QnrS1 structure-activity relationships.
Tavío MM, Jacoby GA, Hooper DC. Tavío MM, et al. J Antimicrob Chemother. 2014 Aug;69(8):2102-9. doi: 10.1093/jac/dku102. Epub 2014 Apr 11. J Antimicrob Chemother. 2014. PMID: 24729602 Free PMC article. - Mutational Analysis of Quinolone Resistance Protein QnrVC7 Provides Novel Insights into the Structure-Activity Relationship of Qnr Proteins.
Po KH, Chan EW, Chen S. Po KH, et al. Antimicrob Agents Chemother. 2016 Jan 11;60(3):1939-42. doi: 10.1128/AAC.01805-15. Antimicrob Agents Chemother. 2016. PMID: 26824937 Free PMC article.
References
- Buchko, G. W., S. Ni, H. Robinson, E. A. Welsh, H. B. Pakrasi, and M. A. Kennedy. 2006. Characterization of two potentially universal turn motifs that shape the repeated five-residues fold-crystal structure of a lumenal pentapeptide repeat protein from Cyanothece 51142. Protein Sci. 15:2579-2595. - PMC - PubMed
- Buchko, G. W., H. Robinson, H. B. Pakrasi, and M. A. Kennedy. 2008. Insights into the structural variation between pentapeptide repeat proteins-crystal structure of Rfr23 from Cyanothece 51142. J. Struct. Biol. 162:184-192. - PubMed
- Cattoir, V., and P. Nordmann. 2009. Plasmid-mediated quinolone resistance in gram-negative bacterial species: an update. Curr. Med. Chem. 16:1028-1046. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BBS/E/J/00000201/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01 AI033696/AI/NIAID NIH HHS/United States
- AI33696/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources