Rapid and transient recruitment of DNMT1 to DNA double-strand breaks is mediated by its interaction with multiple components of the DNA damage response machinery - PubMed (original) (raw)
Rapid and transient recruitment of DNMT1 to DNA double-strand breaks is mediated by its interaction with multiple components of the DNA damage response machinery
Kyungsoo Ha et al. Hum Mol Genet. 2011.
Abstract
DNA methylation is an epigenetic mark critical for regulating transcription, chromatin structure and genome stability. Although many studies have shed light on how methylation impacts transcription and interfaces with the histone code, far less is known about how it regulates genome stability. We and others have shown that DNA methyltransferase 1 (DNMT1), the maintenance methyltransferase, contributes to the cellular response to DNA damage, yet DNMT1's exact role in this process remains unclear. DNA damage, particularly in the form of double-strand breaks (DSBs), poses a major threat to genome integrity. Cells therefore possess a potent system to respond to and repair DSBs, or to initiate cell death. In the current study, we used a near-infrared laser microirradiation system to directly study the link between DNMT1 and DSBs. Our results demonstrate that DNMT1 is rapidly but transiently recruited to DSBs. DNMT1 recruitment is dependent on its ability to interact with both PCNA and the ATR effector kinase CHK1, but is independent of its catalytic activity. In addition, we show for the first time that DNMT1 interacts with the 9-1-1 PCNA-like sliding clamp and that this interaction also contributes to DNMT1 localization to DNA DSBs. Finally, we demonstrate that DNMT1 modulates the rate of DSB repair and is essential for suppressing abnormal activation of the DNA damage response in the absence of exogenous damage. Taken together, our studies provide compelling additional evidence for DNMT1 acting as a regulator of genome integrity and as an early responder to DNA DSBs.
Figures
Figure 1.
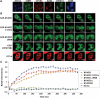
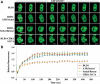
DNMT1 is rapidly recruited to sites of DNA DSBs independent of its catalytic activity. (A) DNA damage response proteins known to be recruited to IRIF (KU80, γH2AX and 53BP1) and the presence of broken DNA ends (shown by TUNEL staining) demonstrate the validity of the NIR laser microirradiation system used in subsequent experiments. DNMT1 is also recruited to NIR laser-irradiated regions. Images are taken 5 min post-NIR laser-induced damage (in a line pattern) using HCT116 cells. (B) Time course of factor recruitment to laser-irradiated regions in live cells. HCT116 cells were transfected with the indicated GFP-tagged DNMT1 or DNMT3 constructs and subjected to NIR laser-induced DNA damage 48 h later. DNMT1 is visible at DSBs within 30 s of laser microirradiation. ΔPBD, deletion of the PCNA-binding domain; C1229A, catalytic center mutation of DNMT1. PCNA, HCT116 cells transfected with RFP-PCNA. Representative images are shown with the irradiated region denoted by the white box and time (in seconds) after irradiation indicated in white text. Bar: 10 µm. (C) Graphical summary of factor recruitment over ∼7.5 min based on the live-cell imaging in (B). At least 10 cells were quantified for each time point (values are the average, and error bars represent the standard deviation). Accumulation curves of fluorescent proteins at sites of damage were obtained by measuring average fluorescence within the laser target area using Image J. Data are presented as the fluorescence level relative to the pre-NIR laser damage level for each construct/transfection.
Figure 2.
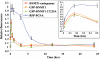
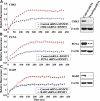
DNMT1 and PCNA are recruited to DSBs with similar kinetics but are lost at different rates. Graphical summary of long-term recruitment/decay kinetics of the indicated DNMT1 constructs compared with PCNA at regions of DSBs. Following transfection of HCT116 cells and NIR laser irradiation 48 h later, cells were fixed at the indicated time points and the degree of fluorescence at regions of DNA damage quantified and set relative to the pre-damage level. Damaged regions were identified by co-staining all slides with γH2AX antibody (not shown). Also shown is a summary of immunostaining for endogenous DNMT1 to demonstrate that GFP-DNMT1 behaves similarly to native DNMT1. DNMT1 is essentially undetectable at DSBs by 3 h post-laser IR, whereas PCNA persists until ∼6–8 h. A blow-up of the early time points is shown in the inset and is similar to the rapid live-cell kinetics in Figure 1C.
Figure 3.
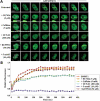
DNMT1 recruitment to regions of DNA damage is reduced by pharmacologic inhibition of the ATR/CHK1 pathway but not by DNA-PKcs or ATM inhibition. (A) HCT116 cells were transfected with wild-type GFP-DNMT1 for 24 h, followed by treatment with the indicated inhibitors for an additional 24 h. Cells were then subjected to NIR laser irradiation and the effect on DNMT1 recruitment to DSBs monitored by live-cell imaging. Representative images are shown. NU7026 preferentially inhibits DNA-PKcs, caffeine inhibits ATM and ATR, KU-55933 inhibits ATM and UCN-01 inhibits CHK1. (B) Graphical summary of DNMT1 recruitment to sites of DNA DSBs and the impact of DNA damage-signaling inhibitors. 5-aza-2′-deoxycytidine (5-azadC) was used as a control and is known to completely block DNMT1 mobility. Bar:10 µm.
Figure 4.
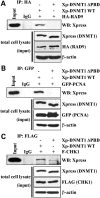
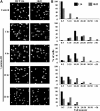
DNMT1 interacts with multiple components of the DNA damage machinery. Wild-type epitope-tagged DNMT1 or DNMT1 lacking the PCNA-binding domain (DNMT1 ΔPBD) was used in co-immunoprecipitation (IP) reactions to examine their ability to interact with (A) RAD9, (B) PCNA and (C) CHK1. The ability of DNMT1 to interact with PCNA and the 9-1-1 (RAD9-RAD1-HUS1) PCNA-like clamp complex subunit RAD9 is dependent on the PBD region; however, the interaction between DNMT1 and CHK1 is not mediated solely by the PBD. Inputs and a loading control (β-actin) are shown below each IP panel. Xp, Xpress epitope tag; IgG, negative control species-matched IP.
Figure 5.
ATR status influences DNMT1 recruitment to regions of DNA DSBs. (A) DLD1 parental or isogenic DLD1 lines expressing a hypomorphic mutant form of ATR (ATR Seckel) or a mutant form of CHK1 that cannot be phosphorylated on the serine 317 position (CHK1 S317A) generated by homologous recombination were used for laser microirradiation studies. DLD1 ATR Seckel cells engineered to re-express WT ATR (Seckel Rescue) were used as a control. Each cell line was transfected with GFP-DNMT1 and laser-irradiated; representative live-cell images are shown. (B) Graphical summary of the effect of each mutant on DNMT1 localization to DSBs. CHK1 S345 was not examined because mutation of this site is incompatible with cell viability.
Figure 6.
CHK1, the 9-1-1 complex and PCNA influence recruitment of DNMT1 to DSBs. (A) GFP-DNMT1 and an shRNA plasmid targeting CHK1 were co-transfected into HCT116 cells. The shRNA plasmid also expressed RFP, enabling microscopic identification of transfected cells (
Supplementary Material, Fig. S1
). Red (shRNA-positive) and green (DNMT1-positive) cells were laser-irradiated and the localization of DNMT monitored by live-cell imaging (
Supplementary Material, Fig. S2
). Results are summarized in graphical form as recruitment versus time (left panel). Western blotting of whole-cell extract from transfected HCT116 cells demonstrates the efficiency of shRNA knockdown (right panel). β-Actin serves as a loading control. Identical studies were performed to monitor GFP-DNMT1 recruitment in cells transfected with (B) PCNA shRNA plasmid and (C) RAD9 shRNA plasmid.
Figure 7.
DNMT1 modulates the DNA damage response in unperturbed and DNA-damaged cells. (A) Western blotting for levels of DNA damage response factors or their activated (phosphorylated) forms in unperturbed HCT116 and HCT116 hypomorphic mutant DNMT1 (1KO) cells. The recently discovered CHK1 kinase target in chromatin, histone H3 threonine 11 (H3T11) (19), was also examined. Total histone H3, DNMT1 and β-actin are shown as controls for loading and identifying each cell line. Chromatin extracts were used for histone protein westerns. HCT116 and 1KO cells were transfected with GFP-DNMT1, subject to DSB generation by laser irradiation, and the recruitment and decay of (B) ATM, ATR (phosphorylated forms), DNMT1, (C) γH2AX and 53BP1 and (D) the replicative and damage clamps PCNA and 9-1-1 (RAD9) were monitored over 24 h. Irradiated cells were fixed at the indicated time points and immunostained. Regions of laser-induced damage were identified by co-staining with γH2AX antibody (not shown). Fluorescence values are set relative to those in the same cell line before DSB induction.
Figure 8.
DNMT1 status influences the rate of DSB repair. (A) Parental and hypomorphic DNMT1 mutant 1KO HCT116 cells before (control) and 1, 6, 12 and 24 h after induction of DNA DSBs with 10 Gy of γ-irradiation (+IR) were used for neutral comet assays. Representative micrographs of cells stained with SYBR green from each treatment are shown. Images are taken at 20× magnification. (B) Quantitation of comet assay data as groups of tail moment scores (an indicator of the level of DNA damage) versus the percentage of cells with each tail moment for each treatment. Fifty randomly selected cells from each treatment were photographed using an Axioplan 3 fluorescent microscope. Tail moment (tail length × percentage of DNA in the tail) was scored by using TriTek Comet Score software.
Similar articles
- DNA methylation inhibitor 5-Aza-2'-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B.
Palii SS, Van Emburgh BO, Sankpal UT, Brown KD, Robertson KD. Palii SS, et al. Mol Cell Biol. 2008 Jan;28(2):752-71. doi: 10.1128/MCB.01799-07. Epub 2007 Nov 8. Mol Cell Biol. 2008. PMID: 17991895 Free PMC article. - Replication independent DNA double-strand break retention may prevent genomic instability.
Kongruttanachok N, Phuangphairoj C, Thongnak A, Ponyeam W, Rattanatanyong P, Pornthanakasem W, Mutirangura A. Kongruttanachok N, et al. Mol Cancer. 2010 Mar 31;9:70. doi: 10.1186/1476-4598-9-70. Mol Cancer. 2010. PMID: 20356374 Free PMC article. - PCNA clamp facilitates action of DNA cytosine methyltransferase 1 on hemimethylated DNA.
Iida T, Suetake I, Tajima S, Morioka H, Ohta S, Obuse C, Tsurimoto T. Iida T, et al. Genes Cells. 2002 Oct;7(10):997-1007. doi: 10.1046/j.1365-2443.2002.00584.x. Genes Cells. 2002. PMID: 12354094 - DNMT1: catalytic and non-catalytic roles in different biological processes.
Mohan KN. Mohan KN. Epigenomics. 2022 May;14(10):629-643. doi: 10.2217/epi-2022-0035. Epub 2022 Apr 12. Epigenomics. 2022. PMID: 35410490 Review. - Maintaining Genome Integrity: Protein Kinases and Phosphatases Orchestrate the Balancing Act of DNA Double-Strand Breaks Repair in Cancer.
Qin S, Kitty I, Hao Y, Zhao F, Kim W. Qin S, et al. Int J Mol Sci. 2023 Jun 16;24(12):10212. doi: 10.3390/ijms241210212. Int J Mol Sci. 2023. PMID: 37373360 Free PMC article. Review.
Cited by
- TET3-mediated DNA oxidation promotes ATR-dependent DNA damage response.
Jiang D, Wei S, Chen F, Zhang Y, Li J. Jiang D, et al. EMBO Rep. 2017 May;18(5):781-796. doi: 10.15252/embr.201643179. Epub 2017 Mar 21. EMBO Rep. 2017. PMID: 28325772 Free PMC article. - The emerging role of epigenetic modifiers in repair of DNA damage associated with chronic inflammatory diseases.
Ding N, Maiuri AR, O'Hagan HM. Ding N, et al. Mutat Res Rev Mutat Res. 2019 Apr-Jun;780:69-81. doi: 10.1016/j.mrrev.2017.09.005. Epub 2017 Sep 28. Mutat Res Rev Mutat Res. 2019. PMID: 31395351 Free PMC article. Review. - Histone and DNA Methylation as Epigenetic Regulators of DNA Damage Repair in Gastric Cancer and Emerging Therapeutic Opportunities.
De Marco K, Sanese P, Simone C, Grossi V. De Marco K, et al. Cancers (Basel). 2023 Oct 13;15(20):4976. doi: 10.3390/cancers15204976. Cancers (Basel). 2023. PMID: 37894343 Free PMC article. Review. - Meta-analysis of DNA double-strand break response kinetics.
Kochan JA, Desclos ECB, Bosch R, Meister L, Vriend LEM, van Attikum H, Krawczyk PM. Kochan JA, et al. Nucleic Acids Res. 2017 Dec 15;45(22):12625-12637. doi: 10.1093/nar/gkx1128. Nucleic Acids Res. 2017. PMID: 29182755 Free PMC article. - Targeted DNA methylation by homology-directed repair in mammalian cells. Transcription reshapes methylation on the repaired gene.
Morano A, Angrisano T, Russo G, Landi R, Pezone A, Bartollino S, Zuchegna C, Babbio F, Bonapace IM, Allen B, Muller MT, Chiariotti L, Gottesman ME, Porcellini A, Avvedimento EV. Morano A, et al. Nucleic Acids Res. 2014 Jan;42(2):804-21. doi: 10.1093/nar/gkt920. Epub 2013 Oct 16. Nucleic Acids Res. 2014. PMID: 24137009 Free PMC article.
References
- Goll M.G., Bestor T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. - PubMed
- Okano M., Bell D.W., Haber D.A., Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous