Cytosolic Ca2+ buffers - PubMed (original) (raw)
Review
Cytosolic Ca2+ buffers
Beat Schwaller. Cold Spring Harb Perspect Biol. 2010 Nov.
Abstract
"Ca(2+) buffers," a class of cytosolic Ca(2+)-binding proteins, act as modulators of short-lived intracellular Ca(2+) signals; they affect both the temporal and spatial aspects of these transient increases in [Ca(2+)](i). Examples of Ca(2+) buffers include parvalbumins (α and β isoforms), calbindin-D9k, calbindin-D28k, and calretinin. Besides their proven Ca(2+) buffer function, some might additionally have Ca(2+) sensor functions. Ca(2+) buffers have to be viewed as one of the components implicated in the precise regulation of Ca(2+) signaling and Ca(2+) homeostasis. Each cell is equipped with proteins, including Ca(2+) channels, transporters, and pumps that, together with the Ca(2+) buffers, shape the intracellular Ca(2+) signals. All of these molecules are not only functionally coupled, but their expression is likely to be regulated in a Ca(2+)-dependent manner to maintain normal Ca(2+) signaling, even in the absence or malfunctioning of one of the components.
Figures
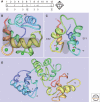
Figure 1.
3D-structures of selected EF-hand Ca2+ buffers. (A) Consensus sequence of the canonical EF-hand Ca2+-binding loop of 12 amino acids. Amino acids X, Y, Z, and –Z provide side-chain oxygen ligands, * provides the backbone carbonyl oxygen, and at –X, a water molecule is hydrogen-bonded to a loop residue. Amino acids most often present at a given position are shown below, and shaded residues are the most conserved ones (Marsden et al. 1990). At positions X and –Z, Asp (D) and Glu (E) are generally present, respectively. The seven oxygen ligands coordinating the Ca2+ ion are located at the seven corners of a pentagonal bipyramid, and the Ca2+ ion (not shown) is in the center (right). (B) Solution structure of Ca2+-bound human α PV; PDB: 1RJV. Both the CD domain (green) and EF domain (yellow/red) bind one Ca2+ ion each (green spheres) in canonical Ca2+-binding loops of 12 amino acids. The orthogonally oriented helices E and F (gray-shaded) are connected by the Ca2+-binding loop. Both Ca2+-binding sites in PV are of the Ca2+/Mg2+ mixed type. The N-terminal AB domain (blue) is necessary for protein stability. (C) NMR solution structure of bovine CB-D9k; PDB: 1B1G. The shown structure takes into account the Ca2+ ions and explicit solvent molecules. The N-terminal domain EF1 is a pseudo (Ψ) EF-hand with a larger loop of 14 amino acids, while the second domain (EF2) has a canonical Ca2+-binding loop of 12 amino acids. In both loops, the Glu residue at the position –Z with the 2 carboxyl oxygen atoms (red) serves as a bidentate ligand representing two corners of the pentagonal bipyramid. This residue, most often Glu (rarely Asp), is a critical determinant for the Ca2+ affinity of the entire loop; Ca2+ ions are shown as green spheres. The two Ca2+-binding loops are in close proximity and stabilized via short β-type interactions (gray-shaded area). (D) 3D NMR structure of CB-D28k; PDB: 2G9B. CB-D28k has a relatively compact structure comprising three Ca2+-binding units, each unit consisting of a pair of EF-hands. Ca2+-binding is restricted to the Ca2+-binding loop 1 in the N-terminal unit (blue), to both loops 3 and 4 in the middle unit (green), and to loop 5 in the C-terminal pair (yellow/red). EF-hands 2 and 6 are nonfunctional, with respect to Ca2+-binding. The Ca2+-binding loops flanked by two almost perpendicular alpha-helical regions are numbered from 1 to 6. Images B–D were generated with PDB ProteinWorkshop 1.50 (Moreland et al. 2005).
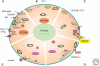
Figure 2.
Homeostatic/adaptive changes in the soma of Purkinje cells (PC) caused by malfunctioning or elimination of Ca2+-signaling toolkit component(s); regulation by the Ca2+ homeostasome. (A) A detailed situation is depicted for wild-type mice. Increases in [Ca2+]i (red arrows) result from influx via CaV2.1 (P/Q) channels or release from internal stores (light blue) via the IP3 receptor. IP3 is generated by the activation of metabotropic glutamate receptors (mGluR). Ca2+ removal systems (blue arrows) include PMCA and NCX in the plasma membrane, SERCA pumps, and mitochondria (green). Identified Ca2+-signaling toolkit components including organelles, which are up- or down-regulated, are marked in yellow and magenta, respectively. (B) In PV−/−, PC subplasmalemmal mitochondria are increased, while ER volume directly underneath the plasma membrane is decreased. (C) In addition to the changes observed in PV−/−, in PC lacking both, PV and CB-D28k the auxiliary Cavβ2a subunit of CaV2.1 (P/Q), is decreased, leading to increased voltage-dependent inactivation of P-type currents. Model studies indicate an up-regulation of Ca2+ extrusion systems, possibly PMCA. (D) In PMCA2−/− PC, expression of mGluR1 and of IP3 receptor type 1 (IP3R1), responsible for the Ca2+ release from ER stores, is decreased. Also, the cytosolic Ca2+ buffering capacity mediated by CB-D28k is decreased. (E) In leaner mice PC that are characterized by strongly attenuated Cav2.1 Ca2+ channel function, the rapid Ca2+ buffering/sequestering capacity is reduced: PV and CB-D28k are down-regulated and (subplasmalemmal) ER is decreased/impaired, leading to reduced Ca2+ uptake.
Similar articles
- Cytosolic Ca2+ Buffers Are Inherently Ca2+ Signal Modulators.
Schwaller B. Schwaller B. Cold Spring Harb Perspect Biol. 2020 Jan 2;12(1):a035543. doi: 10.1101/cshperspect.a035543. Cold Spring Harb Perspect Biol. 2020. PMID: 31308146 Free PMC article. Review. - The continuing disappearance of "pure" Ca2+ buffers.
Schwaller B. Schwaller B. Cell Mol Life Sci. 2009 Jan;66(2):275-300. doi: 10.1007/s00018-008-8564-6. Cell Mol Life Sci. 2009. PMID: 19099190 Free PMC article. Review. - The use of transgenic mouse models to reveal the functions of Ca2+ buffer proteins in excitable cells.
Schwaller B. Schwaller B. Biochim Biophys Acta. 2012 Aug;1820(8):1294-303. doi: 10.1016/j.bbagen.2011.11.008. Epub 2011 Nov 27. Biochim Biophys Acta. 2012. PMID: 22138448 Review. - Calcium buffering properties of calbindin D28k and parvalbumin in rat sensory neurones.
Chard PS, Bleakman D, Christakos S, Fullmer CS, Miller RJ. Chard PS, et al. J Physiol. 1993 Dec;472:341-57. doi: 10.1113/jphysiol.1993.sp019950. J Physiol. 1993. PMID: 8145149 Free PMC article. - Distribution of calretinin, calbindin D28k, and parvalbumin in subcellular fractions of rat cerebellum: effects of calcium.
Winsky L, Kuźnicki J. Winsky L, et al. J Neurochem. 1995 Jul;65(1):381-8. doi: 10.1046/j.1471-4159.1995.65010381.x. J Neurochem. 1995. PMID: 7790883
Cited by
- Calcium, Bioenergetics, and Parkinson's Disease.
Zampese E, Surmeier DJ. Zampese E, et al. Cells. 2020 Sep 8;9(9):2045. doi: 10.3390/cells9092045. Cells. 2020. PMID: 32911641 Free PMC article. Review. - ROS Homeostasis in Abiotic Stress Tolerance in Plants.
Nadarajah KK. Nadarajah KK. Int J Mol Sci. 2020 Jul 23;21(15):5208. doi: 10.3390/ijms21155208. Int J Mol Sci. 2020. PMID: 32717820 Free PMC article. Review. - A novel explanation for observed CaMKII dynamics in dendritic spines with added EGTA or BAPTA.
Matolcsi M, Giordano N. Matolcsi M, et al. Biophys J. 2015 Feb 17;108(4):975-985. doi: 10.1016/j.bpj.2014.12.044. Biophys J. 2015. PMID: 25692602 Free PMC article. - Absence of the calcium-binding protein calretinin, not of calbindin D-28k, causes a permanent impairment of murine adult hippocampal neurogenesis.
Todkar K, Scotti AL, Schwaller B. Todkar K, et al. Front Mol Neurosci. 2012 Apr 23;5:56. doi: 10.3389/fnmol.2012.00056. eCollection 2012. Front Mol Neurosci. 2012. PMID: 22536174 Free PMC article. - Nanodomain coupling between Ca²⁺ channels and sensors of exocytosis at fast mammalian synapses.
Eggermann E, Bucurenciu I, Goswami SP, Jonas P. Eggermann E, et al. Nat Rev Neurosci. 2011 Dec 20;13(1):7-21. doi: 10.1038/nrn3125. Nat Rev Neurosci. 2011. PMID: 22183436 Free PMC article. Review.
References
- Airaksinen L, Virkkala J, Aarnisalo A, Meyer M, Ylikoski J, Airaksinen MS 2000. Lack of calbindin-D28k does not affect hearing level or survival of hair cells in acoustic trauma. ORL J Otorhinolaryngol Relat Spec 62: 9–12 - PubMed
- Akke M, Forsen S, Chazin WJ 1991. Molecular basis for co-operativity in Ca2+ binding to calbindin D9k. 1H nuclear magnetic resonance studies of (Cd2+)1-bovine calbindin D9k. J Mol Biol 220: 173–189 - PubMed
- Allbritton NL, Meyer T, Stryer L 1992. Range of messenger action of calcium ion and inositol 1,4,5,-trisphophate. Science 258: 1812–1815 - PubMed
- Babini E, Bertini I, Capozzi F, Del Bianco C, Hollender D, Kiss T, Luchinat C, Quattrone A 2004. Solution structure of human beta-parvalbumin and structural comparison with its paralog alpha-parvalbumin and with their rat orthologs. Biochemistry 43: 16076–16085 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous