Macroscale spatial variation in chronic wound microbiota: a cross-sectional study - PubMed (original) (raw)
. 2011 Jan-Feb;19(1):80-8.
doi: 10.1111/j.1524-475X.2010.00628.x. Epub 2010 Oct 13.
Cindy M Liu, Yelena M Frankel, Johan H Melendez, Maliha Aziz, Jordan Buchhagen, Tania Contente-Cuomo, David M Engelthaler, Paul S Keim, Jacques Ravel, Gerald S Lazarus, Jonathan M Zenilman
Affiliations
- PMID: 20946140
- PMCID: PMC3022109
- DOI: 10.1111/j.1524-475X.2010.00628.x
Macroscale spatial variation in chronic wound microbiota: a cross-sectional study
Lance B Price et al. Wound Repair Regen. 2011 Jan-Feb.
Abstract
Controlling for sample site is considered to be an important aspect of chronic wound microbiological investigations; yet, macroscale spatial variation in wound microbiota has not been well characterized. A total of 31 curette samples were collected at the leading edge, opposing leading edge, and/or center of 13 chronic wounds. Bacterial community composition was characterized using a combination of 16S rRNA gene-based pyrosequencing; heat map display; hierarchical clustering; nonmetric multidimensional scaling; and permutation multivariate analysis of variance. A total of 58 bacterial families and 91 bacterial genera were characterized among the 13 wounds. While substantial macroscale spatial variation was observed among the wounds, bacterial communities at different sites within individual wounds were significantly more similar than those in different wounds (p=0.001). Our results support the prevalent opinion that controlling for sample site may improve the quality of wound microbiota studies; however, the significant similarity in bacterial communities from different sites within individual wounds indicates that studies failing to control for sampling site should not be disregarded based solely on this criterion. A composite sample from multiple sites across the surface of individual wounds may provide the most robust characterization of wound microbiota.
© 2010 by the Wound Healing Society.
Figures
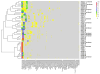
Figure 1
Figure 1A. Heat map display of bacterial families comprising chronic wound microbiota (samples are grouped based on hierarchical clustering of bacterial communities). The color key for the number of sequences from each bacterial family is shown on the right. A = leading edge; B = apposing leading edge; C = center; Combined = simulated combined wound; TR01 and TR02 (1, 2, & 3) are technical replicates created by dividing a single curette sample into three equal parts prior to processing. Figure 1B. Heat map display of bacterial genera comprising chronic wound microbiota (samples are grouped based on hierarchical clustering of bacterial communities). The color key for the number of sequences from each bacterial family is shown on the right. (A = leading edge; B = apposing leading edge; C = center; Combined = simulated combined wound; TR01 and TR02 (1, 2, & 3) are technical replicates).
Figure 1
Figure 1A. Heat map display of bacterial families comprising chronic wound microbiota (samples are grouped based on hierarchical clustering of bacterial communities). The color key for the number of sequences from each bacterial family is shown on the right. A = leading edge; B = apposing leading edge; C = center; Combined = simulated combined wound; TR01 and TR02 (1, 2, & 3) are technical replicates created by dividing a single curette sample into three equal parts prior to processing. Figure 1B. Heat map display of bacterial genera comprising chronic wound microbiota (samples are grouped based on hierarchical clustering of bacterial communities). The color key for the number of sequences from each bacterial family is shown on the right. (A = leading edge; B = apposing leading edge; C = center; Combined = simulated combined wound; TR01 and TR02 (1, 2, & 3) are technical replicates).
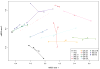
Figure 2
Non-metric Multi-Dimensional Scaling (nMDS) plot of microbiota of wounds samples taken from different sites within the wounds. (A = leading edge; B = apposing leading edge; C = center; star = simulated combined wound; TR01 and TR02 (1, 2, & 3) are technical replicates).
Similar articles
- Community analysis of chronic wound bacteria using 16S rRNA gene-based pyrosequencing: impact of diabetes and antibiotics on chronic wound microbiota.
Price LB, Liu CM, Melendez JH, Frankel YM, Engelthaler D, Aziz M, Bowers J, Rattray R, Ravel J, Kingsley C, Keim PS, Lazarus GS, Zenilman JM. Price LB, et al. PLoS One. 2009 Jul 31;4(7):e6462. doi: 10.1371/journal.pone.0006462. PLoS One. 2009. PMID: 19649281 Free PMC article. - Bacteria Detected in both Urine and Open Wounds in Nursing Home Residents: a Pilot Study.
Libertucci J, Bassis CM, Cassone M, Gibson K, Lansing B, Mody L, Young VB, Meddings J. Libertucci J, et al. mSphere. 2019 Aug 28;4(4):e00463-19. doi: 10.1128/mSphere.00463-19. mSphere. 2019. PMID: 31462413 Free PMC article. - Analysis of the chronic wound microbiota of 2,963 patients by 16S rDNA pyrosequencing.
Wolcott RD, Hanson JD, Rees EJ, Koenig LD, Phillips CD, Wolcott RA, Cox SB, White JS. Wolcott RD, et al. Wound Repair Regen. 2016 Jan-Feb;24(1):163-74. doi: 10.1111/wrr.12370. Epub 2015 Dec 10. Wound Repair Regen. 2016. PMID: 26463872 - The Impact of Microbial Communities on Wound Healing: A Review.
Xu Z, Hsia HC. Xu Z, et al. Ann Plast Surg. 2018 Jul;81(1):113-123. doi: 10.1097/SAP.0000000000001450. Ann Plast Surg. 2018. PMID: 29746280 Review. - Bacterial Contribution in Chronicity of Wounds.
Rahim K, Saleha S, Zhu X, Huo L, Basit A, Franco OL. Rahim K, et al. Microb Ecol. 2017 Apr;73(3):710-721. doi: 10.1007/s00248-016-0867-9. Epub 2016 Oct 14. Microb Ecol. 2017. PMID: 27742997 Review.
Cited by
- The microbiome in wound repair and tissue fibrosis.
Scales BS, Huffnagle GB. Scales BS, et al. J Pathol. 2013 Jan;229(2):323-31. doi: 10.1002/path.4118. Epub 2012 Nov 29. J Pathol. 2013. PMID: 23042513 Free PMC article. Review. - Association between baseline abundance of Peptoniphilus, a Gram-positive anaerobic coccus, and wound healing outcomes of DFUs.
Min KR, Galvis A, Baquerizo Nole KL, Sinha R, Clarke J, Kirsner RS, Ajdic D. Min KR, et al. PLoS One. 2020 Jan 24;15(1):e0227006. doi: 10.1371/journal.pone.0227006. eCollection 2020. PLoS One. 2020. PMID: 31978071 Free PMC article. - Successful topical treatment of human biofilms using multiple antibiotic elution from a collagen-rich hydrogel.
Sharma AD, Jarman EH, Kuppalli K, Murphy MJ, Longaker MT, Gurtner G, Fox PM. Sharma AD, et al. Sci Rep. 2024 Mar 7;14(1):5621. doi: 10.1038/s41598-024-54477-z. Sci Rep. 2024. PMID: 38454046 Free PMC article. - Rapid analysis of metagenomic data using signature-based clustering.
Chappell T, Geva S, Hogan JM, Huygens F, Rathnayake IU, Rudd S, Kelly W, Perrin D. Chappell T, et al. BMC Bioinformatics. 2018 Dec 21;19(Suppl 20):509. doi: 10.1186/s12859-018-2540-4. BMC Bioinformatics. 2018. PMID: 30577803 Free PMC article. - A systematic literature review of the human skin microbiome as biomarker for dermatological drug development.
Niemeyer-van der Kolk T, van der Wall HEC, Balmforth C, Van Doorn MBA, Rissmann R. Niemeyer-van der Kolk T, et al. Br J Clin Pharmacol. 2018 Oct;84(10):2178-2193. doi: 10.1111/bcp.13662. Epub 2018 Jul 19. Br J Clin Pharmacol. 2018. PMID: 29877593 Free PMC article.
References
- Mustoe TA, O'Shaughnessy K, Kloeters O. Chronic wound pathogenesis and current treatment strategies: A unifying hypothesis. Plast Reconstr Surg. 2006;117(7 Suppl):35S–41S. - PubMed
- Fonder MA, Lazarus GS, Cowan DA, Aronson-Cook B, Kohli AR, Mamelak AJ. Treating the chronic wound: A practical approach to the care of nonhealing wounds and wound care dressings. J Am Acad Dermatol. 2008;158:185–206. - PubMed
- Andersen A, Hill KE, Stephens P, Thomas DW, Jorgensen B, Krogfelt KA. Bacterial profiling using skin grafting, standard culture and molecular bacteriological methods. J Wound Care. 2007;16:171–5. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical