A comprehensive survey of clonal diversity measures in Barrett's esophagus as biomarkers of progression to esophageal adenocarcinoma - PubMed (original) (raw)
A comprehensive survey of clonal diversity measures in Barrett's esophagus as biomarkers of progression to esophageal adenocarcinoma
Lauren M F Merlo et al. Cancer Prev Res (Phila). 2010 Nov.
Abstract
Neoplastic progression is an evolutionary process driven by the generation of clonal diversity and natural selection on that diversity within a neoplasm. We hypothesized that clonal diversity is associated with risk of progression to cancer. We obtained molecular data from a cohort of 239 participants with Barrett's esophagus, including microsatellite shifts and loss of heterozygosity, DNA content tetraploidy and aneuploidy, methylation, and sequence mutations. Using these data, we tested all major diversity measurement methods, including genetic divergence and entropy-based measures, to determine which measures are correlated with risk of progression to esophageal adenocarcinoma. We also tested whether the use of different sets of loci and alterations to define clones (e.g., selectively advantageous versus evolutionarily neutral) improved the predictive value of the diversity indices. All diversity measures were strong and highly significant predictors of progression (Cox proportional hazards model, P < 0.001). The type of alterations evaluated had little effect on the predictive value of most of the diversity measures. In summary, diversity measures are robust predictors of progression to cancer in this cohort.
©2010 AACR.
Figures
Figure 1
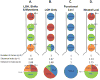
Example of effect of the distribution of clones on diversity indices for different Hill numbers (values of q) (27). Participants 1 and 2 have 4 clones (Diversity index=4 at q =0). In this hypothetical example, as q increases, a single dominant clone in Participants 2 and 3 has an increasingly strong negative effect on the total diversity. In other words, the rare clones in Particpants 2 and 3 have less impact on diversity as q increases. The addition of 3 more rare clones in Participant 3, relative to Participant 2, has a large impact on the q =0 diversity measure (number of clones), a small effect on the Shannon Index (q =1), and negligible effect on the q =2 (Simpson index) or _q_=3 diversity measures in Participant 3.
Figure 2
Map of the clones in the BE segment from a single participant from our cohort under different definitions of a clone. The pie charts below each segment represent total % of each clone in the BE segment. Defining clones in different ways alters the distribution and number of clones and the diversity indices. A. Both LOH and changes in microsatellite lengths (shifts) in all 18 microsatellites, as well as sequence mutations in CDKN2A (p16/INK4A) and TP53 (p53), and DNA content are used to define a clone. B. Only LOH lesions in the 18 microsatellites and DNA content are used to define a clone, while shifts and mutations are ignored. C. Only lesions that increase the fitness of a clone are used to define a clone. These lesions include LOH or sequence mutations in CDKN2A or TP53, or hypermethylation of the CDKN2A promoter (20). D. DNA content and lesions that have no detectable effect on the fitness of a clone were used, including shifts in any of the 19 microsatellites and LOH on the q arms of chromosomes 9 and 17 (20). This illustrative patient was chosen because the different definitions of clones lead to different diversity measures, though this was often not the case for other participants in the cohort.
Figure 3
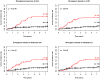
Kaplan-Meier cancer incidence curves for mean pairwise genetic divergence based on four different definitions of clones (p<0.001 in all cases). Red=upper quartile, Black=bottom three quartiles. The (number of cancers/total number of participants) in the upper and lower 3 quartiles are given as numbers to right of the curves.
Figure 4
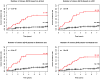
Kaplan-Meier cancer incidence curves for q=0 based on four different definitions of clones (p<0.001 in all cases). Red=upper quartile, Black=bottom three quartiles. The (number of cancers/total number of participants) in the upper and lower 3 quartiles are given as numbers to right of the curves.
Figure 5
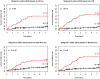
Kaplan-Meier cancer incidence curves for q=2 based on four different definitions of clones (p<0.001 in all cases). ). Red=upper quartile, Black=bottom three quartiles. The (number of cancers/total number of participants) in the upper and lower 3 quartiles are given as numbers to right of the curves.
Comment in
- The origins and implications of intratumor heterogeneity.
Michor F, Polyak K. Michor F, et al. Cancer Prev Res (Phila). 2010 Nov;3(11):1361-4. doi: 10.1158/1940-6207.CAPR-10-0234. Epub 2010 Oct 19. Cancer Prev Res (Phila). 2010. PMID: 20959519 Free PMC article.
Similar articles
- The combination of genetic instability and clonal expansion predicts progression to esophageal adenocarcinoma.
Maley CC, Galipeau PC, Li X, Sanchez CA, Paulson TG, Blount PL, Reid BJ. Maley CC, et al. Cancer Res. 2004 Oct 15;64(20):7629-33. doi: 10.1158/0008-5472.CAN-04-1738. Cancer Res. 2004. PMID: 15492292 - Clonal expansion and loss of heterozygosity at chromosomes 9p and 17p in premalignant esophageal (Barrett's) tissue.
Galipeau PC, Prevo LJ, Sanchez CA, Longton GM, Reid BJ. Galipeau PC, et al. J Natl Cancer Inst. 1999 Dec 15;91(24):2087-95. doi: 10.1093/jnci/91.24.2087. J Natl Cancer Inst. 1999. PMID: 10601379 Free PMC article. - Genetic and Epigenetic Alterations in Barrett's Esophagus and Esophageal Adenocarcinoma.
Kaz AM, Grady WM, Stachler MD, Bass AJ. Kaz AM, et al. Gastroenterol Clin North Am. 2015 Jun;44(2):473-89. doi: 10.1016/j.gtc.2015.02.015. Epub 2015 Apr 1. Gastroenterol Clin North Am. 2015. PMID: 26021206 Free PMC article. Review. - Loss of heterozygosity on chromosome 17p predicts neoplastic progression in Barrett's esophagus.
Dolan K, Morris AI, Gosney JR, Field JK, Sutton R. Dolan K, et al. J Gastroenterol Hepatol. 2003 Jun;18(6):683-9. doi: 10.1046/j.1440-1746.2003.03048.x. J Gastroenterol Hepatol. 2003. PMID: 12753151 - Cellular origins and molecular mechanisms of Barrett's esophagus and esophageal adenocarcinoma.
Fang Y, Chen X, Bajpai M, Verma A, Das KM, Souza RF, Garman KS, Donohoe CL, O'Farrell NJ, Reynolds JV, Dvorak K. Fang Y, et al. Ann N Y Acad Sci. 2013 Oct;1300:187-199. doi: 10.1111/nyas.12249. Ann N Y Acad Sci. 2013. PMID: 24117642 Review.
Cited by
- The origins and implications of intratumor heterogeneity.
Michor F, Polyak K. Michor F, et al. Cancer Prev Res (Phila). 2010 Nov;3(11):1361-4. doi: 10.1158/1940-6207.CAPR-10-0234. Epub 2010 Oct 19. Cancer Prev Res (Phila). 2010. PMID: 20959519 Free PMC article. - An Integrative Analysis of Nasopharyngeal Carcinoma Genomes Unraveled Unique Processes Driving a Viral-Positive Cancer.
Liu X, Li Y, Zhou X, Zhu S, Kaya NA, Chan YS, Ma L, Xu M, Zhai W. Liu X, et al. Cancers (Basel). 2023 Feb 15;15(4):1243. doi: 10.3390/cancers15041243. Cancers (Basel). 2023. PMID: 36831585 Free PMC article. - MAGI1 Prevents Senescence and Promotes the DNA Damage Response in ER+ Breast Cancer.
Wörthmüller J, Disler S, Pradervand S, Richard F, Haerri L, Ruiz Buendía GA, Fournier N, Desmedt C, Rüegg C. Wörthmüller J, et al. Cells. 2023 Jul 25;12(15):1929. doi: 10.3390/cells12151929. Cells. 2023. PMID: 37566008 Free PMC article. - Intratumor and Intertumor Heterogeneity in Melanoma.
Grzywa TM, Paskal W, Włodarski PK. Grzywa TM, et al. Transl Oncol. 2017 Dec;10(6):956-975. doi: 10.1016/j.tranon.2017.09.007. Epub 2017 Oct 24. Transl Oncol. 2017. PMID: 29078205 Free PMC article. Review. - Whole genome sequencing for lung cancer.
Daniels M, Goh F, Wright CM, Sriram KB, Relan V, Clarke BE, Duhig EE, Bowman RV, Yang IA, Fong KM. Daniels M, et al. J Thorac Dis. 2012 Apr 1;4(2):155-63. doi: 10.3978/j.issn.2072-1439.2012.02.01. J Thorac Dis. 2012. PMID: 22833821 Free PMC article.
References
- Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8. - PubMed
- Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6:924–35. - PubMed
- Martin CM, Kehoe L, Spillane CO, O'Leary JJ. Gene discovery in cervical cancer : towards diagnostic and therapeutic biomarkers. Mol Diagn Ther. 2007;11:277–90. - PubMed
- Tabor MP, Braakhuis BJ, van der Wal JE, et al. Comparative molecular and histological grading of epithelial dysplasia of the oral cavity and the oropharynx. J Pathol. 2003;199:354–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 CA132450/CA/NCI NIH HHS/United States
- P30 CA010815/CA/NCI NIH HHS/United States
- P01 CA091955/CA/NCI NIH HHS/United States
- R01 CA107028/CA/NCI NIH HHS/United States
- R03 CA137811/CA/NCI NIH HHS/United States
- R01 CA14065/CA/NCI NIH HHS/United States
- P01 CA91955/CA/NCI NIH HHS/United States
- R01 CA140657/CA/NCI NIH HHS/United States
- R01 CA119224/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical