Ultra-deep sequencing reveals the microRNA expression pattern of the human stomach - PubMed (original) (raw)
. 2010 Oct 8;5(10):e13205.
doi: 10.1371/journal.pone.0013205.
André S Khayat, Artur Silva, Dayse O Alencar, Jessé Lobato, Larissa Luz, Daniel G Pinheiro, Leonardo Varuzza, Monica Assumpção, Paulo Assumpção, Sidney Santos, Dalila L Zanette, Wilson A Silva Jr, Rommel Burbano, Sylvain Darnet
Affiliations
- PMID: 20949028
- PMCID: PMC2951895
- DOI: 10.1371/journal.pone.0013205
Ultra-deep sequencing reveals the microRNA expression pattern of the human stomach
Ândrea Ribeiro-dos-Santos et al. PLoS One. 2010.
Abstract
Background: While microRNAs (miRNAs) play important roles in tissue differentiation and in maintaining basal physiology, little is known about the miRNA expression levels in stomach tissue. Alterations in the miRNA profile can lead to cell deregulation, which can induce neoplasia.
Methodology/principal findings: A small RNA library of stomach tissue was sequenced using high-throughput SOLiD sequencing technology. We obtained 261,274 quality reads with perfect matches to the human miRnome, and 42% of known miRNAs were identified. Digital Gene Expression profiling (DGE) was performed based on read abundance and showed that fifteen miRNAs were highly expressed in gastric tissue. Subsequently, the expression of these miRNAs was validated in 10 healthy individuals by RT-PCR showed a significant correlation of 83.97% (P<0.05). Six miRNAs showed a low variable pattern of expression (miR-29b, miR-29c, miR-19b, miR-31, miR-148a, miR-451) and could be considered part of the expression pattern of the healthy gastric tissue.
Conclusions/significance: This study aimed to validate normal miRNA profiles of human gastric tissue to establish a reference profile for healthy individuals. Determining the regulatory processes acting in the stomach will be important in the fight against gastric cancer, which is the second-leading cause of cancer mortality worldwide.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
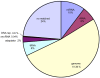
Figure 1. Distribution of small RNAs sequenced from human gastric cardia tissue using deep-sequencing.
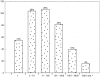
Figure 2. Distribution of microRNA by read count number in human gastric cardia.
The read count is based on quantity of read detected during the deep-sequencing of small RNA library of gastric cardia, using SOLiD system. For microRNA detection was used the miRBase release 15.0.
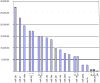
Figure 3. Most expressed microRNAs in human gastric cardia.
The read count is based on quantity of read detected during the deep-sequencing of small RNA library of gastric cardia, using SOLiD system.
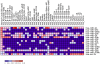
Figure 4. Heatmap of normalized expression of the 15 most expressed mature miRNAs in human gastric tissue and their comparison with other normal tissues published in the mammalian microRNA expression atlas (Landgraf, P. et al. (2007). Cell 129: 1401–1414).
The heatmap was generated using gene pattern software with normalized expression of microRNA (read count number for specific microRNA/total of count number of microRNA). Color scale indicates the percent of total count reads number 0 to 0.2 (0 to 20%). Human tissue description and abbreviation: hsa_B-cell-CD19 (B cells from peripheral blood); hsa_B-cell-CD19-pool (B cells from peripheral blood (pool from 4 healthy donors)); hsa_B-cell-CD19-2 (B cells from peripheral blood); hsa_DC-unstim (myeloid dendritic cells not stimulated); hsa_DC-stim (myeloid dendritic cells stimulated with endotoxin); hsa_DC-unstim (myeloid dendritic cells not stimulated); hsa_DC-stim (myeloid dendritic cells stimulated with endotoxin); hsa_Fibrobl-CMV(Foreskin fibroblasts; Primary fibroblasts lytically infected with Cytomegalovirus); hsa_Frontal-cortex-adult; (Brain_normal adult; sample from Brodman area 9 (superior frontal gyrus) of a 20 year old healthy male, 6 hours postmortem); hsa_Granulocytes-CD1; (Granulocytes; Granulocyte cells from peripheral blood (pool from 4 healthy donors)); hsa_HSC-CD34 (pluripotent hematopoetic stem cell CD34+ - sorted cells); hsa_NK-CD56;(NK cells from peripheral blood (pool from 4 healthy donors)); hsa_Podocytes-Moins-undiff (Podocytes_undifferentiated); hsa_Podocytes-Moins-diff (Podocytes_differentiated); hsa_T-cell-CD4 (T helper cells, peripheral blood (pool from 4 healthy donors)); hsa_T-cell-CD4-2 (T helper cells); hsa_T-cell-CD4-naive(CD4+ CD45 RA+(CD45RO−) native cells); hsa_T-cell-CD4-effector (CD4+, CD45RO+, CD27−, CCR7−. Effector cells); hsa_T-cell-CD4-memory (CD4+CD45RO+(CD45RA−) memory cells); hsa_T-cell-CD8-2 (cytotoxic T-cells); hsa_T-cell-CD8 (cytotoxic T-cells, peripheral blood(pool from 4 healthy donors); hsa_T-cell-CD8-naive (CD8+, CD45RA+, CD27+, CCR7+); hsa_USSC (unrestricted somatic stem cells from umbilical cord); hsa_USSC-d1(unrestricted somatic stem cells from umbilical cord induced 1 day to osteoblasts); hsa_USSC-d3 (unrestricted somatic stem cells from umbilical cord induced 3 day to osteoblasts); hsa_USSC-d7 (unrestricted somatic stem cells from umbilical cord induced 7 day to osteoblasts).
Figure 5. Quantification by Real Time PCR of high expressed microRNAs in human gastric cardia.
The quantification is based on Ct and was normalized by endogenous expression control. The 2−ΔCt for each miR is the mean of ten determination originated from gastric cardia tissue of ten different individuals.
Similar articles
- MiRNA expression profile for the human gastric antrum region using ultra-deep sequencing.
Moreira FC, Assumpção M, Hamoy IG, Darnet S, Burbano R, Khayat A, Gonçalves AN, Alencar DO, Cruz A, Magalhães L, Araújo W Jr, Silva A, Santos S, Demachki S, Assumpção P, Ribeiro-dos-Santos A. Moreira FC, et al. PLoS One. 2014 Mar 19;9(3):e92300. doi: 10.1371/journal.pone.0092300. eCollection 2014. PLoS One. 2014. PMID: 24647245 Free PMC article. - Profiling microRNA expression in bovine alveolar macrophages using RNA-seq.
Vegh P, Foroushani AB, Magee DA, McCabe MS, Browne JA, Nalpas NC, Conlon KM, Gordon SV, Bradley DG, MacHugh DE, Lynn DJ. Vegh P, et al. Vet Immunol Immunopathol. 2013 Oct 1;155(4):238-44. doi: 10.1016/j.vetimm.2013.08.004. Epub 2013 Aug 24. Vet Immunol Immunopathol. 2013. PMID: 24021155 - MicroRNA Expression Profiling to Identify and Validate Reference Genes for the Relative Quantification of microRNA in Rectal Cancer.
Eriksen AH, Andersen RF, Pallisgaard N, Sørensen FB, Jakobsen A, Hansen TF. Eriksen AH, et al. PLoS One. 2016 Mar 3;11(3):e0150593. doi: 10.1371/journal.pone.0150593. eCollection 2016. PLoS One. 2016. PMID: 26937645 Free PMC article. - Comparison of stomach microRNA transcriptomes of Tibetan and Yorkshire pigs by deep sequencing.
Sun WK, Li Y, Cheng C, Chen YH, Zeng K, Chen X, Gu Y, Liu R, Lv X, Gao R. Sun WK, et al. Genes Genomics. 2018 Sep;40(9):937-943. doi: 10.1007/s13258-018-0696-y. Epub 2018 May 14. Genes Genomics. 2018. PMID: 30155707 - Profiling of 95 microRNAs in pancreatic cancer cell lines and surgical specimens by real-time PCR analysis.
Zhang Y, Li M, Wang H, Fisher WE, Lin PH, Yao Q, Chen C. Zhang Y, et al. World J Surg. 2009 Apr;33(4):698-709. doi: 10.1007/s00268-008-9833-0. World J Surg. 2009. PMID: 19030927 Free PMC article. Review.
Cited by
- Loss of Tyrosine Phosphatase Delta Promotes Gastric Cancer Progression via Signal Transducer and Activator of Transcription 3 Pathways.
Wu L, Gao L, Kong D, Xue H. Wu L, et al. Dig Dis Sci. 2019 Nov;64(11):3164-3172. doi: 10.1007/s10620-019-05637-z. Epub 2019 Apr 30. Dig Dis Sci. 2019. PMID: 31041642 - Helicobacter pylori infection: host immune response, implications on gene expression and microRNAs.
Cadamuro AC, Rossi AF, Maniezzo NM, Silva AE. Cadamuro AC, et al. World J Gastroenterol. 2014 Feb 14;20(6):1424-37. doi: 10.3748/wjg.v20.i6.1424. World J Gastroenterol. 2014. PMID: 24587619 Free PMC article. Review. - MicroRNA-148a is downregulated in gastric cancer, targets MMP7, and indicates tumor invasiveness and poor prognosis.
Sakamoto N, Naito Y, Oue N, Sentani K, Uraoka N, Oo HZ, Yanagihara K, Aoyagi K, Sasaki H, Yasui W. Sakamoto N, et al. Cancer Sci. 2014 Feb;105(2):236-43. doi: 10.1111/cas.12330. Epub 2014 Jan 6. Cancer Sci. 2014. PMID: 24283384 Free PMC article. - TargetCompare: A web interface to compare simultaneous miRNAs targets.
Moreira FC, Dustan B, Hamoy IG, Ribeiro-Dos-Santos AM, Dos Santos AR. Moreira FC, et al. Bioinformation. 2014 Sep 30;10(9):602-5. doi: 10.6026/97320630010602. eCollection 2014. Bioinformation. 2014. PMID: 25352731 Free PMC article. - The Role of Mir-148a in Cancer.
Li Y, Deng X, Zeng X, Peng X. Li Y, et al. J Cancer. 2016 Jun 21;7(10):1233-41. doi: 10.7150/jca.14616. eCollection 2016. J Cancer. 2016. PMID: 27390598 Free PMC article. Review.
References
- Ricarte Filho JC, Kimura ET. MicroRNAs: novel class of gene regulators involved in endocrine function and cancer. Arq Bras Endocrinol Metabol. 2006;50:1102–1107. - PubMed
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
- Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38:323–332. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources