Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor - PubMed (original) (raw)
Comparative Study
Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor
Nadja Neumann et al. PLoS One. 2010.
Abstract
Background: The Nuclear Pore Complex (NPC) facilitates molecular trafficking between nucleus and cytoplasm and is an integral feature of the eukaryote cell. It exhibits eight-fold rotational symmetry and is comprised of approximately 30 nucleoporins (Nups) in different stoichiometries. Nups are broadly conserved between yeast, vertebrates and plants, but few have been identified among other major eukaryotic groups.
Methodology/principal findings: We screened for Nups across 60 eukaryote genomes and report that 19 Nups (spanning all major protein subcomplexes) are found in all eukaryote supergroups represented in our study (Opisthokonts, Amoebozoa, Viridiplantae, Chromalveolates and Excavates). Based on parsimony, between 23 and 26 of 31 Nups can be placed in LECA. Notably, they include central components of the anchoring system (Ndc1 and Gp210) indicating that the anchoring system did not evolve by convergence, as has previously been suggested. These results significantly extend earlier results and, importantly, unambiguously place a fully-fledged NPC in LECA. We also test the proposal that transmembrane Pom proteins in vertebrates and yeasts may account for their variant forms of mitosis (open mitoses in vertebrates, closed among yeasts). The distribution of homologues of vertebrate Pom121 and yeast Pom152 is not consistent with this suggestion, but the distribution of fungal Pom34 fits a scenario wherein it was integral to the evolution of closed mitosis in ascomycetes. We also report an updated screen for vesicle coating complexes, which share a common evolutionary origin with Nups, and can be traced back to LECA. Surprisingly, we find only three supergroup-level differences (one gain and two losses) between the constituents of COPI, COPII and Clathrin complexes.
Conclusions/significance: Our results indicate that all major protein subcomplexes in the Nuclear Pore Complex are traceable to the Last Eukaryotic Common Ancestor (LECA). In contrast to previous screens, we demonstrate that our conclusions hold regardless of the position of the root of the eukaryote tree.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
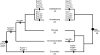
Figure 1. NPC structure, composition and Nup conservation across eukaryotes.
a) Schematic section through the nuclear pore complex. Sub-complexes are indicated as boxes and marked in different colors to indicate their position in the pore. b) The table summarizes Nup distribution across eukaryotic super-groups. Color-coding matches that of the subcomplexes in (a). Nucleoporins indicated with bold letters are universally distributed across eukaryotes, as judged by presence in at least one genome from each of the five supergroups.
Figure 2. NPC components are traceable to LECA.
NPC pore composition in LECA based on two alternative rootings of the eukaryote tree. In the left hand tree, Excavates are the outgroup. The right hand tree is rooted on the basis of the unikont/bikont bifurcation. Gains (+) and losses (–) in different lineages are indicated under each scenario. Where gains and losses are equally probable, these are marked with (?).
Similar articles
- Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex.
Mans BJ, Anantharaman V, Aravind L, Koonin EV. Mans BJ, et al. Cell Cycle. 2004 Dec;3(12):1612-37. doi: 10.4161/cc.3.12.1316. Epub 2004 Dec 20. Cell Cycle. 2004. PMID: 15611647 - Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor.
DeGrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, Rout MP, Chait BT. DeGrasse JA, et al. Mol Cell Proteomics. 2009 Sep;8(9):2119-30. doi: 10.1074/mcp.M900038-MCP200. Epub 2009 Jun 13. Mol Cell Proteomics. 2009. PMID: 19525551 Free PMC article. - AI-based structure prediction empowers integrative structural analysis of human nuclear pores.
Mosalaganti S, Obarska-Kosinska A, Siggel M, Taniguchi R, Turoňová B, Zimmerli CE, Buczak K, Schmidt FH, Margiotta E, Mackmull MT, Hagen WJH, Hummer G, Kosinski J, Beck M. Mosalaganti S, et al. Science. 2022 Jun 10;376(6598):eabm9506. doi: 10.1126/science.abm9506. Epub 2022 Jun 10. Science. 2022. PMID: 35679397 - Evolution: functional evolution of nuclear structure.
Wilson KL, Dawson SC. Wilson KL, et al. J Cell Biol. 2011 Oct 17;195(2):171-81. doi: 10.1083/jcb.201103171. J Cell Biol. 2011. PMID: 22006947 Free PMC article. Review. - [Nuclear pores: from yeast to higher eukaryotes].
Doye V. Doye V. J Soc Biol. 2002;196(4):349-54. J Soc Biol. 2002. PMID: 12645306 Review. French.
Cited by
- Molecular basis for Nup37 and ELY5/ELYS recruitment to the nuclear pore complex.
Bilokapic S, Schwartz TU. Bilokapic S, et al. Proc Natl Acad Sci U S A. 2012 Sep 18;109(38):15241-6. doi: 10.1073/pnas.1205151109. Epub 2012 Sep 5. Proc Natl Acad Sci U S A. 2012. PMID: 22955883 Free PMC article. - An introduction to dynamic nucleoporins in Leishmania species: Novel targets for tropical-therapeutics.
Dubey AK, Kumar P, Mandal D, Ravichandiran V, Singh SK. Dubey AK, et al. J Parasit Dis. 2022 Dec;46(4):1176-1191. doi: 10.1007/s12639-022-01515-0. Epub 2022 Jul 25. J Parasit Dis. 2022. PMID: 36457769 Free PMC article. Review. - Structural evolution of the membrane-coating module of the nuclear pore complex.
Liu X, Mitchell JM, Wozniak RW, Blobel G, Fan J. Liu X, et al. Proc Natl Acad Sci U S A. 2012 Oct 9;109(41):16498-503. doi: 10.1073/pnas.1214557109. Epub 2012 Sep 26. Proc Natl Acad Sci U S A. 2012. PMID: 23019579 Free PMC article. - Manifold Routes to a Nucleus.
Hendrickson HL, Poole AM. Hendrickson HL, et al. Front Microbiol. 2018 Oct 26;9:2604. doi: 10.3389/fmicb.2018.02604. eCollection 2018. Front Microbiol. 2018. PMID: 30416499 Free PMC article. - The Ancestral Mitotic State: Closed Orthomitosis With Intranuclear Spindles in the Syncytial Last Eukaryotic Common Ancestor.
Bremer N, Tria FDK, Skejo J, Martin WF. Bremer N, et al. Genome Biol Evol. 2023 Mar 3;15(3):evad016. doi: 10.1093/gbe/evad016. Genome Biol Evol. 2023. PMID: 36752808 Free PMC article.
References
- Alcazar-Roman AR, Tran EJ, Guo S, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol. 2006;8:711–716. - PubMed
- Tran EJ, Wente SR. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–1053. - PubMed
- Rout MP, Aitchison JD. Pore relations: nuclear pore complexes and nucleocytoplasmic exchange. Essays Biochem. 2000;36:75–88. - PubMed
- Lim RY, Aebi U, Stoffler D. From the trap to the basket: getting to the bottom of the nuclear pore complex. Chromosoma. 2006;115:15–26. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases