Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap - PubMed (original) (raw)
Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap
Ricardo M Neto-Silva et al. Dev Cell. 2010.
Abstract
An understanding of how animal size is controlled requires knowledge of how positive and negative growth regulatory signals are balanced and integrated within cells. Here we demonstrate that the activities of the conserved growth-promoting transcription factor Myc and the tumor-suppressing Hippo pathway are codependent during growth of Drosophila imaginal discs. We find that Yorkie (Yki), the Drosophila homolog of the Hippo pathway transducer, Yap, regulates the transcription of Myc, and that Myc functions as a critical cellular growth effector of the pathway. We demonstrate that in turn, Myc regulates the expression of Yki as a function of its own cellular level, such that high levels of Myc repress Yki expression through both transcriptional and posttranscriptional mechanisms. We propose that the codependent regulatory relationship functionally coordinates the cellular activities of Yki and Myc and provides a mechanism of growth control that regulates organ size and has broad implications for cancer.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures
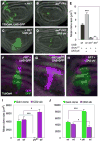
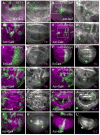
Figure 1. Yki requires dMyc for growth and local differences in Hpo activity induce cell competition
(A–E) Loss of dMyc prevents Yki-induced growth. MARCM clones generated in wing discs, marked by T80Gal4>UAS-GFP expression (green). (A) control clones; (B) dm4 mutant clones; (C) UAS-_yki_-expressing clones; (D) dm4 mutant clones that express UAS-yki. (E) Quantification of mean clone size. UAS-_yki_-expressing clones (grey bar) induce significant growth relative to control clones (black bar), but its expression fails to induce growth in dm4 mutant cells (dark grey bar) even when death is prevented by UAS-droncCD expression (white bar). Error bars in this and all subsequent figures are standard error of the mean (SEM). (F–J) Measurement of cell competition using MARCM in wing discs to generate a Gal4/UAS-_GFP_-expressing clone (green) and its sibling clone (marked with 2 copies of CD2, magenta). (F) Control TubGal4>UAS-GFP clone and wildtype (wt) CD2 sibling showing that wt sibling clones grow equally well; (G) ykiB5 mutant TubGal4>UAS-GFP clone and wt CD2 sibling; (H) UAS-_yki_-expressing TubGal4>UAS-GFP clone and wt CD2 sibling. (I, J) Quantification of cell competition. (I) ykiB5 mutant TubGal4>UAS-GFP clones grow less than control wt TubGal4>UAS-GFP clones (green bars) and are competed against by their wt and heterozygous neighbors, while their wt CD2 sibling clones grow significantly more than control CD2 sibling clones (magenta bars), an indication of “winner” status. (J) UAS-_yki_-expressing TubGal4>UAS-GFP clones grow more than control TubGal4>UAS-GFP clones (green bars), while their wt sibling CD2 clones are smaller than control wt sibling CD2 clones (magenta bars), indicative of “loser” status. See also Figure S1. P-values, * p < 0.05; *** p < 0.001. In this and all subsequent figures, discs are oriented with dorsal up and posterior to the right.
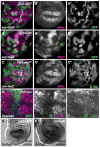
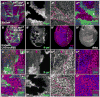
Figure 2. Hpo signaling activity regulates dMyc expression
(A, A′) Control wing disc with ActGal4>GFP-expressing clones (green) showing wildtype dMyc expression (magenta) (A′). (B, B′) Clonal expression of UAS-yki (green) up-regulates dMyc expression (magenta) in endogenous regions and induces it ectopically. Note that clones at the dorso-ventral boundary (arrowhead in B′) do not express dMyc due to dominant patterning cues (Johnston et al 1999). (C, C′) Clones of wtsx1 mutant cells (absence of green) induce expression of dMyc. (D–D″) Wing disc expressing UAS-hpo and UAS-p35 under DppGal4 control (green). dMyc is specifically down-regulated by UAS-hpo expression (arrowhead, D′), whereas Nub expression (blue), which is expressed throughout the wing pouch, is unaffected (arrowhead, D″). See also Figure S2. (E–F) dmyc mRNA in control wing disc (E) or wing disc that expresses UAS-yki in posterior cells with EnGal4 (right side of dotted line in F). dmyc mRNA is increased in posterior cells in response to UAS-yki expression (F).
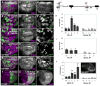
Figure 3. dMyc is a transcriptional target of the Hpo pathway
(A–A″) Wing disc expressing UAS-GFP, UAS-dcr-2 and a UAS- short-hairpin (sh) sd under DppGal4 control (green). Reduction of sd expression reduces dMyc expression in the wing pouch (arrow, A′), but Nub expression is generally unaffected (A″). (B–B″) Clones expressing UAS-GFP (B″) and UAS-yki upregulate dMyc (magenta) in the wing pouch and ectopically activate dMyc in proximal cells (arrow in B′, B″). Arrowhead points to a clone at the D–V boundary that does not alter dMyc expression. (C–C″) GFP-marked clones expressing UAS-yki, UAS-sh_sd_, and UAS-dcr-2. Reduced Sd expression prevents dMyc upregulation (magenta) in its endogenous domain and ectopic activation in proximal cells (arrow in C′, C″). (D–E) Beta-galactosidase (β-gal) expression (magenta) from a P-element insertion (G0359) into the proximal promoter region of the dm locus. See also Figure S3. (D–D″) Control wing disc. (E–E″) Wing disc expressing UAS-yki in clones. G0359-lacZ responds strongly to UAS-yki expression. GFP marked clones are shown in E″. (F) β-gal expression from a P-element insertion (PL35) into intron 2 of the dm locus. (G, G′) Expression of Yki under Engal4 control leads to strong upregulation of β-gal (magenta). (H–J) Schematic representation of the dm locus. Black rectangles are coding regions, white rectangles are non-coding regions; lines denote introns. Red bars indicate consensus Sd binding sites and green lines are degenerate Sd binding sites in the locus; red triangles represent Yki-responsive P-element insertions. Blocks labeled A–F represent regions amplified in the chIP experiments; Pr, Promoter; In2, Intron2. (H) Yki chIP on the dm locus. Anti-Yki (Yki IP) or IgG (mock IP) antibodies were used to precipitate chromatin from wt discs; quantitative RT-PCR was done on regions A–D and on a negative control locus, pyruvate dehydrogenase (PD). Graph shows the enrichment of signal over input for A, B, C, D and PD. Amplicons B, C and D were all consistently enriched over PD in each of 4 independent experiments, although statistical significance was obtained only for amplicon B, due to noise between experiments. (I) ChIP data covering amplicon E, in the first coding exon (a negative control) and amplicon F, covering a single Sd site. Neither amplicon was enriched relative to PD in the experiments. (J) Sd-GFP chIP on the dm locus. Amplicons B, C, and D showed significant enrichment after precipitation of wt disc chromatin with antibodies against GFP. The strongest enrichment occurred of amplicon D, where a cluster of three highly conserved Sd sites is located (not shown;
). Inset shows expression from Sd-GFP line. P-values, ** p<0.01.
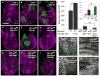
Figure 4. Co-dependence of dMyc and Yki in growth regulation
(A–I) MARCM TubGal4>UAS-GFP clones (green) that express UAS-dmyc (alone or together with other transgenes) in yki mutant clones; their wt CD2 sibling clones are magenta. (A) Discs with control clones; (B) clones expressing UAS-dmyc; (C) clones expressing UAS-yki; (D) ykiB5 mutant clones; (E) ykiB5 mutant clones rescued by yki expression; (F) ykiB5 mutant clones that express UAS-dmyc; (G) ykiB5 mutant clones that express UAS-diap1*; (H) ykiB5 mutant clones that co-express UAS-dmyc and UAS-diap1*; (I) ykiB5 mutant clones that co-express UAS-dmyc and UAS-bantam. (J) Clone size quantification. UAS-diap1* or UAS-bantam expression slightly rescues growth of ykiB5 clones. UAS-dmyc significantly increases size of wt clones but fails to increase ykiB5 mutant clone size, even with co-expression of UAS-diap1* or UAS-bantam. (K–N) Fibrillarin staining in wing imaginal discs with clones expressing dmyc (K); with ykiB5 mutant clones expressing dmyc (L); with clones expressing dmyc + bantam (M); and with ykiB5 mutant clones expressing UAS-dmyc + UAS-bantam. UAS-dmyc upregulates Fibrillarin and increases nucleolar size in wt cells (K) and in ykiB5 mutant cells (L, N), indicating that some growth-promoting targets of dMyc are independent of Yki. Green lines in K–N indicate clone boundaries. See also Figure S4. P-value, *** p<0.001.
Figure 5. dMyc regulates Yki expression and activity
Panels A–H show Yki immunostaining (magenta) in wing discs. (A, A′) Control wing disc clones; (B, B′) and dm4 mutant clones. Clones are marked by the absence of green (arm-lacZ, dotted line indicates clone boundaries). B′, high magnification of area boxed in B. dm4 clones have elevated Yki expression (arrows). (C–D) Control ActGal4>UAS-GFP clones (green) (C, C′) or clones expressing UAS-sh_dmyc_ (D, D′). D′ is a higher magnification of area boxed in D. Moderate reduction of dMyc is sufficient to increase Yki protein (arrows). (E–E′) Wing discs expressing UAS-dmyc (E, E′) or UAS-sh_dmyc_ (F, F′) in posterior cells (green) under EnGal4 control. Yki expression is decreased in high dMyc cells throughout the disc, and increased in low dMyc cells within the wing pouch (upper and lower boundaries are marked by arrows in F′). See also Figure S5. (G, G′) Clones expressing dmyc, marked with UAS-GFP expression (green). Yki is decreased in clones expressing dmyc (arrow). (H–H′) Clones expressing dmyc in a ykiB5 animal rescued with a Tub-yki transgene. All Yki expression is from the transgene. UAS-dmyc represses Tub-yki in the clones (arrows), indicating that dMyc also regulates Yki expression post-transcriptionally. dmyc expression also reduces yki mRNA 30% below wildtype levels (see Figure S3). (I–L) Expression of Yki targets is diminished by UAS-dmyc expression. dIAP1 protein (I), expanded-lacZ (J, J′), Four Jointed (K, K′), and fj-lacZ (L, L′) are all downregulated (arrows) in clones expressing UAS-dmyc.
Figure 6. Wildtype levels of dMyc regulate Yki
(A) Wing disc with wts_x1mutant clones immunostained for Yki (red). (A′) Close-up view of the region boxed in (A). wtsx1 mutant cells are marked by absence of β-gal. (A″) Yki expression. In the wts mutant clone Yki is somewhat diffuse, with both nuclear and cytoplasmic localization. (A‴) Close-up of merged channels of box in (A). (B) Yki staining (red) in wing disc with wts mutant clones and expressing UAS-sh_dmyc in posterior cells under EnGal4 control. wts mutant cells are marked by lack of β-gal (B′). Posterior cells are located to the right of the dashed yellow line (B′, B″) and show reduced levels of dMyc (B‴). Box A is located in the anterior compartment and box P in the posterior. (C–C‴) Close up view of the region inside box P. The red dotted line marks the wts_x1clone boundary, mutant cells lack β-gal. Cells to the right of the yellow dashed line express UAS-sh_dmyc; the arrow points to non-mutant tissue (β-gal-positive). Note the substantial accumulation of Yki in the nucleus of wts_x1 cells that have reduced dMyc levels due to expression of UAS-sh_dmyc. (D–D″) Close up view of the region inside box A. _wts_x1 mutant clone (red line in D) located in the anterior compartment is marked by the absence of β-gal (D′). As in A″, Yki staining is more diffuse, with some nuclear localization (D″), but substantially less than in _wts_x1 clones located in the posterior compartment, which express lower levels of dMyc (compare to C″).
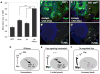
Figure 7. A model for a homeostatic feedback mechanism that prevents runaway growth
(A) Size of Act>Gal4 cell clones expressing UAS-GFP, UAS-dmyc, UAS-yki or co-expressing UAS-yki and UAS-dmyc. Simultaneous expression of Yki and dMyc drives signficantly more growth than expression of either factor alone. *** p < 0.001**. (B–C)** Cell death, assessed by TUNEL staining (red), in wing discs with wildtype (B) or wtsx1 mutant clones (C) where UAS-dmyc was expressed in the posterior compartment. wtsx1 cells are marked by absence of β-gal (green) and the posterior compartment is marked by absence of Ci, a marker of anterior cells (blue). wtsx1 mutant tissue is protected from cell death induced by de-regulated dMyc (arrows). (D–F) A model of co-dependent regulation between Yki and dMyc. In wt cells (D), nuclear (active) Yki activates expression of dmyc and other Hpo target genes (diap1, bantam, fj). Negative feedback between dMyc activity and Yki expression keeps the activity of each in balance. (E) In wts mutant cells, Hpo signaling is inactivated; more active Yki increases dMyc expression. High dMyc expression and activity will decrease Yki expression, which in turn will curb dMyc expression, resetting a balance and limiting over-growth. (F) Cells expressing deregulated levels of dMyc will decrease Yki expression, resulting in loss of Yki pro-survival targets and cell death. In the model, deregulation of both dMyc and Yki expression will abolish the negative feedback loop and promote overgrowth; in mammals, this could lead to cancer.
Similar articles
- Growth control: Myc and Yorkie get connected.
Stocker H. Stocker H. Curr Biol. 2011 Jan 11;21(1):R37-9. doi: 10.1016/j.cub.2010.11.057. Curr Biol. 2011. PMID: 21215937 - Scaling the Drosophila Wing: TOR-Dependent Target Gene Access by the Hippo Pathway Transducer Yorkie.
Parker J, Struhl G. Parker J, et al. PLoS Biol. 2015 Oct 16;13(10):e1002274. doi: 10.1371/journal.pbio.1002274. eCollection 2015 Oct. PLoS Biol. 2015. PMID: 26474042 Free PMC article. - Mechanical strain regulates the Hippo pathway in Drosophila.
Fletcher GC, Diaz-de-la-Loza MD, Borreguero-Muñoz N, Holder M, Aguilar-Aragon M, Thompson BJ. Fletcher GC, et al. Development. 2018 Mar 8;145(5):dev159467. doi: 10.1242/dev.159467. Development. 2018. PMID: 29440303 Free PMC article. - A role for Hipk in the Hippo pathway.
Heidary Arash E, Attisano L. Heidary Arash E, et al. Sci Signal. 2013 May 14;6(275):pe18. doi: 10.1126/scisignal.2004259. Sci Signal. 2013. PMID: 23674821 Review. - Yorkie: the final destination of Hippo signaling.
Oh H, Irvine KD. Oh H, et al. Trends Cell Biol. 2010 Jul;20(7):410-7. doi: 10.1016/j.tcb.2010.04.005. Epub 2010 May 7. Trends Cell Biol. 2010. PMID: 20452772 Free PMC article. Review.
Cited by
- TAZ expression as a prognostic indicator in colorectal cancer.
Yuen HF, McCrudden CM, Huang YH, Tham JM, Zhang X, Zeng Q, Zhang SD, Hong W. Yuen HF, et al. PLoS One. 2013;8(1):e54211. doi: 10.1371/journal.pone.0054211. Epub 2013 Jan 23. PLoS One. 2013. PMID: 23372686 Free PMC article. - The Initial Stage of Tumorigenesis in Drosophila Epithelial Tissues.
Tamori Y. Tamori Y. Adv Exp Med Biol. 2019;1167:87-103. doi: 10.1007/978-3-030-23629-8_5. Adv Exp Med Biol. 2019. PMID: 31520350 Review. - Cell competition and its implications for development and cancer.
Tamori Y, Deng WM. Tamori Y, et al. J Genet Genomics. 2011 Oct 20;38(10):483-95. doi: 10.1016/j.jgg.2011.09.006. Epub 2011 Sep 21. J Genet Genomics. 2011. PMID: 22035869 Free PMC article. Review. - Cellular sensory mechanisms for detecting specific fold-changes in extracellular cues.
Hironaka K, Morishita Y. Hironaka K, et al. Biophys J. 2014 Jan 7;106(1):279-88. doi: 10.1016/j.bpj.2013.10.039. Biophys J. 2014. PMID: 24411260 Free PMC article. - Ribosomal protein mutations and cell competition: autonomous and nonautonomous effects on a stress response.
Kiparaki M, Baker NE. Kiparaki M, et al. Genetics. 2023 Jul 6;224(3):iyad080. doi: 10.1093/genetics/iyad080. Genetics. 2023. PMID: 37267156 Free PMC article.
References
- Badouel C, Gardano L, Amin N, Garg A, Rosenfeld R, Le Bihan T, McNeill H. The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev Cell. 2009a;16:411–420. - PubMed
- Badouel C, Garg A, McNeill H. Herding Hippos: regulating growth in flies and man. Curr Opin Cell Biol 2009b - PubMed
- Bourbon HM, Gonzy-Treboul G, Peronnet F, Alin MF, Ardourel C, Benassayag C, Cribbs D, Deutsch J, Ferrer P, Haenlin M, et al. A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech Dev. 2002;110:71–83. - PubMed
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases