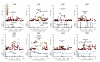A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1 - PubMed (original) (raw)
. 2010 Nov;42(11):985-90.
doi: 10.1038/ng.694. Epub 2010 Oct 17.
Genetic Analysis of Psoriasis Consortium & the Wellcome Trust Case Control Consortium 2 1; Francesca Capon, Chris C A Spencer, Jo Knight, Michael E Weale, Michael H Allen, Anne Barton, Gavin Band, Céline Bellenguez, Judith G M Bergboer, Jenefer M Blackwell, Elvira Bramon, Suzannah J Bumpstead, Juan P Casas, Michael J Cork, Aiden Corvin, Panos Deloukas, Alexander Dilthey, Audrey Duncanson, Sarah Edkins, Xavier Estivill, Oliver Fitzgerald, Colin Freeman, Emiliano Giardina, Emma Gray, Angelika Hofer, Ulrike Hüffmeier, Sarah E Hunt, Alan D Irvine, Janusz Jankowski, Brian Kirby, Cordelia Langford, Jesús Lascorz, Joyce Leman, Stephen Leslie, Lotus Mallbris, Hugh S Markus, Christopher G Mathew, W H Irwin McLean, Ross McManus, Rotraut Mössner, Loukas Moutsianas, Asa T Naluai, Frank O Nestle, Giuseppe Novelli, Alexandros Onoufriadis, Colin N A Palmer, Carlo Perricone, Matti Pirinen, Robert Plomin, Simon C Potter, Ramon M Pujol, Anna Rautanen, Eva Riveira-Munoz, Anthony W Ryan, Wolfgang Salmhofer, Lena Samuelsson, Stephen J Sawcer, Joost Schalkwijk, Catherine H Smith, Mona Ståhle, Zhan Su, Rachid Tazi-Ahnini, Heiko Traupe, Ananth C Viswanathan, Richard B Warren, Wolfgang Weger, Katarina Wolk, Nicholas Wood, Jane Worthington, Helen S Young, Patrick L J M Zeeuwen, Adrian Hayday, A David Burden, Christopher E M Griffiths, Juha Kere, André Reis, Gilean McVean, David M Evans, Matthew A Brown, Jonathan N Barker, Leena Peltonen, Peter Donnelly, Richard C Trembath
Affiliations
- PMID: 20953190
- PMCID: PMC3749730
- DOI: 10.1038/ng.694
A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1
Genetic Analysis of Psoriasis Consortium & the Wellcome Trust Case Control Consortium 2 et al. Nat Genet. 2010 Nov.
Abstract
To identify new susceptibility loci for psoriasis, we undertook a genome-wide association study of 594,224 SNPs in 2,622 individuals with psoriasis and 5,667 controls. We identified associations at eight previously unreported genomic loci. Seven loci harbored genes with recognized immune functions (IL28RA, REL, IFIH1, ERAP1, TRAF3IP2, NFKBIA and TYK2). These associations were replicated in 9,079 European samples (six loci with a combined P < 5 × 10⁻⁸ and two loci with a combined P < 5 × 10⁻⁷). We also report compelling evidence for an interaction between the HLA-C and ERAP1 loci (combined P = 6.95 × 10⁻⁶). ERAP1 plays an important role in MHC class I peptide processing. ERAP1 variants only influenced psoriasis susceptibility in individuals carrying the HLA-C risk allele. Our findings implicate pathways that integrate epidermal barrier dysfunction with innate and adaptive immune dysregulation in psoriasis pathogenesis.
Figures
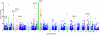
Figure 1
Plot of genome wide association results. Genome wide association results from 523,067 SNPs on chromosomes 1-22 and 12,408 SNPs on the X chromosome using the additive model in SNPTEST. The −log10 (P) values are thresholded at 10−10. Regions in red are described in table 2. Regions which have been shown previously to be associated with psoriasis and which replicate in this study are highlighted in green, as described in table 1.
Figure 2
Regional association plots. The -log10 (P) values for SNPs at eight new loci are shown on the upper part of each plot. SNPs are coloured based on their _r_2 with the labeled hit SNP which has the smallest P value in the region. _r_2 is calculated from the 58C control data. Where more than one SNP is labeled, there is evidence for multiple signals at the locus (see text). The bottom section of each plot shows the fine scale recombination rates estimated from HapMap individuals and genes are marked by horizontal blue lines. Genes within the recombination region of the hit SNP are labeled except for 19p13, where the genes are: GLP-1, FDX1L, RAVER1, ICAM3, TKY2, CDC37, PDE4A, KEAP1, S1PR5.
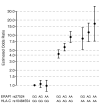
Figure 3
Statistical interaction between ERAP1 and HLA-C genotypes. Odds ratio estimates for ERAP1 genotypes at rs27524 stratified by HLA-C genotypes at rs10484554. Odds ratios were estimated by fitting a logistic regression model which included the first principal component as a covariate and a parameter for each of the possible two-locus genotypes. Note that odds are measured relative to the most protective two-locus genotype (which by definition has an odds ratio of 1). Bars represent 95% confidence intervals calculated from the standard error of the estimates of the genotype parameters in logistic regression model.
Similar articles
- Paediatric-onset psoriasis is associated with ERAP1 and IL23R loci, LCE3C_LCE3B deletion and HLA-C*06.
Bergboer JG, Oostveen AM, de Jager ME, den Heijer M, Joosten I, van de Kerkhof PC, Zeeuwen PL, de Jong EM, Schalkwijk J, Seyger MM. Bergboer JG, et al. Br J Dermatol. 2012 Oct;167(4):922-5. doi: 10.1111/j.1365-2133.2012.10992.x. Br J Dermatol. 2012. PMID: 22512642 - The association of ERAP1 and ERAP2 single nucleotide polymorphisms and their haplotypes with psoriasis vulgaris is dependent on the presence or absence of the HLA-C*06:02 allele and age at disease onset.
Wiśniewski A, Matusiak Ł, Szczerkowska-Dobosz A, Nowak I, Łuszczek W, Kuśnierczyk P. Wiśniewski A, et al. Hum Immunol. 2018 Feb;79(2):109-116. doi: 10.1016/j.humimm.2017.11.010. Epub 2017 Nov 26. Hum Immunol. 2018. PMID: 29183862 - Genetic association with ERAP1 in psoriasis is confined to disease onset after puberty and not dependent on HLA-C*06.
Lysell J, Padyukov L, Kockum I, Nikamo P, Ståhle M. Lysell J, et al. J Invest Dermatol. 2013 Feb;133(2):411-7. doi: 10.1038/jid.2012.280. Epub 2012 Aug 30. J Invest Dermatol. 2013. PMID: 22931917 Free PMC article. - The genetics of psoriasis and psoriatic arthritis.
Chandran V. Chandran V. Clin Rev Allergy Immunol. 2013 Apr;44(2):149-56. doi: 10.1007/s12016-012-8303-5. Clin Rev Allergy Immunol. 2013. PMID: 22274791 Review. - The role of polymorphic ERAP1 in autoinflammatory disease.
Reeves E, James E. Reeves E, et al. Biosci Rep. 2018 Aug 29;38(4):BSR20171503. doi: 10.1042/BSR20171503. Print 2018 Sep 3. Biosci Rep. 2018. PMID: 30054427 Free PMC article. Review.
Cited by
- The genetic history of Greenlandic-European contact.
Waples RK, Hauptmann AL, Seiding I, Jørsboe E, Jørgensen ME, Grarup N, Andersen MK, Larsen CVL, Bjerregaard P, Hellenthal G, Hansen T, Albrechtsen A, Moltke I. Waples RK, et al. Curr Biol. 2021 May 24;31(10):2214-2219.e4. doi: 10.1016/j.cub.2021.02.041. Epub 2021 Mar 11. Curr Biol. 2021. PMID: 33711251 Free PMC article. - Endoplasmic reticulum aminopeptidase 1 and rheumatic disease: functional variation.
Tran TM, Colbert RA. Tran TM, et al. Curr Opin Rheumatol. 2015 Jul;27(4):357-63. doi: 10.1097/BOR.0000000000000188. Curr Opin Rheumatol. 2015. PMID: 26002027 Free PMC article. Review. - From genetics of inflammatory bowel disease towards mechanistic insights.
Graham DB, Xavier RJ. Graham DB, et al. Trends Immunol. 2013 Aug;34(8):371-8. doi: 10.1016/j.it.2013.04.001. Epub 2013 Apr 30. Trends Immunol. 2013. PMID: 23639549 Free PMC article. Review. - Dense genotyping of immune-related susceptibility loci reveals new insights into the genetics of psoriatic arthritis.
Bowes J, Budu-Aggrey A, Huffmeier U, Uebe S, Steel K, Hebert HL, Wallace C, Massey J, Bruce IN, Bluett J, Feletar M, Morgan AW, Marzo-Ortega H, Donohoe G, Morris DW, Helliwell P, Ryan AW, Kane D, Warren RB, Korendowych E, Alenius GM, Giardina E, Packham J, McManus R, FitzGerald O, McHugh N, Brown MA, Ho P, Behrens F, Burkhardt H, Reis A, Barton A. Bowes J, et al. Nat Commun. 2015 Feb 5;6:6046. doi: 10.1038/ncomms7046. Nat Commun. 2015. PMID: 25651891 Free PMC article. - Enhanced meta-analysis and replication studies identify five new psoriasis susceptibility loci.
Tsoi LC, Spain SL, Ellinghaus E, Stuart PE, Capon F, Knight J, Tejasvi T, Kang HM, Allen MH, Lambert S, Stoll SW, Weidinger S, Gudjonsson JE, Koks S, Kingo K, Esko T, Das S, Metspalu A, Weichenthal M, Enerback C, Krueger GG, Voorhees JJ, Chandran V, Rosen CF, Rahman P, Gladman DD, Reis A, Nair RP, Franke A, Barker JNWN, Abecasis GR, Trembath RC, Elder JT. Tsoi LC, et al. Nat Commun. 2015 May 5;6:7001. doi: 10.1038/ncomms8001. Nat Commun. 2015. PMID: 25939698 Free PMC article.
References
- Nestle FO, Kaplan DH, Barker J. Psoriasis. N Engl J Med. 2009;361:496–509. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G0700314/MRC_/Medical Research Council/United Kingdom
- 085780/WT_/Wellcome Trust/United Kingdom
- 083948/Z/07/Z/WT_/Wellcome Trust/United Kingdom
- 068545/Z/02/WT_/Wellcome Trust/United Kingdom
- PDA/02/06/016/DH_/Department of Health/United Kingdom
- G0800582/MRC_/Medical Research Council/United Kingdom
- G0901310/MRC_/Medical Research Council/United Kingdom
- G0601387/MRC_/Medical Research Council/United Kingdom
- G0000934/MRC_/Medical Research Council/United Kingdom
- G19/2/MRC_/Medical Research Council/United Kingdom
- 079643/WT_/Wellcome Trust/United Kingdom
- G0600705/MRC_/Medical Research Council/United Kingdom
- 084726/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous