Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies - PubMed (original) (raw)
Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies
Sébastien Boisvert et al. J Comput Biol. 2010 Nov.
Abstract
An accurate genome sequence of a desired species is now a pre-requisite for genome research. An important step in obtaining a high-quality genome sequence is to correctly assemble short reads into longer sequences accurately representing contiguous genomic regions. Current sequencing technologies continue to offer increases in throughput, and corresponding reductions in cost and time. Unfortunately, the benefit of obtaining a large number of reads is complicated by sequencing errors, with different biases being observed with each platform. Although software are available to assemble reads for each individual system, no procedure has been proposed for high-quality simultaneous assembly based on reads from a mix of different technologies. In this paper, we describe a parallel short-read assembler, called Ray, which has been developed to assemble reads obtained from a combination of sequencing platforms. We compared its performance to other assemblers on simulated and real datasets. We used a combination of Roche/454 and Illumina reads to assemble three different genomes. We showed that mixing sequencing technologies systematically reduces the number of contigs and the number of errors. Because of its open nature, this new tool will hopefully serve as a basis to develop an assembler that can be of universal utilization (availability: http://deNovoAssembler.sf.Net/). For online Supplementary Material , see www.liebertonline.com.
Figures
FIG. 1.
A subgraph of a de Bruijn graph. This figure shows a part of a de Bruijn graph. In this example, short reads are not enough for the assembly problem. Suppose that the true genome sequence is of the form . If the length of the reads (or paired reads) is smaller than the
subsequence, no hints will help an assembly algorithm to differentiate the true sequence from the following one
. On the other hand, if there is a read that starts before _z_1 and ends after _z_2, there will be a possibility to solve this branching problem.
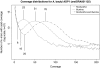
FIG. 2.
Coverage distributions. This figure shows the coverage distributions of _k_-mers for the A. baylyi ADP1 dataset with Roche/454, Illumina, and Roche/454 and Illumina, k = 21. The minimum coverage and the peak coverage are identified for the Roche/454, Illumina, and Roche/454 and Illumina coverage distributions. The peak coverage of Roche/454+Illumina is greater than the sum of the peak coverage of Roche/454 and the peak coverage of Illumina, which suggests that the mixed approach allows one to recover low-coverage regions.
FIG. 3.
The Ray algorithm. Ray is a greedy algorithm on a de Bruijn graph. The extension of seeds is carried out by the subroutine GrowSeed. Each seed is extended using the set of Rules 1 and 2. Afterwards, each extended seed is extended in the opposite direction using the reverse-complement path of the extended seed. Given two seeds _s_1 and _s_2, the reachability of _s_1 from _s_2 is not a necessary and sufficient condition of the reachability of _s_2 from _s_1. Owing to this property of reachability between seeds, a final merging step is necessary to remove things appearing twice in the assembly.
Similar articles
- Subset selection of high-depth next generation sequencing reads for de novo genome assembly using MapReduce framework.
Fang CH, Chang YJ, Chung WC, Hsieh PH, Lin CY, Ho JM. Fang CH, et al. BMC Genomics. 2015;16 Suppl 12(Suppl 12):S9. doi: 10.1186/1471-2164-16-S12-S9. Epub 2015 Dec 9. BMC Genomics. 2015. PMID: 26678408 Free PMC article. - QuorUM: An Error Corrector for Illumina Reads.
Marçais G, Yorke JA, Zimin A. Marçais G, et al. PLoS One. 2015 Jun 17;10(6):e0130821. doi: 10.1371/journal.pone.0130821. eCollection 2015. PLoS One. 2015. PMID: 26083032 Free PMC article. - Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads.
Wick RR, Judd LM, Gorrie CL, Holt KE. Wick RR, et al. PLoS Comput Biol. 2017 Jun 8;13(6):e1005595. doi: 10.1371/journal.pcbi.1005595. eCollection 2017 Jun. PLoS Comput Biol. 2017. PMID: 28594827 Free PMC article. - Sequence assembly using next generation sequencing data--challenges and solutions.
Chin FY, Leung HC, Yiu SM. Chin FY, et al. Sci China Life Sci. 2014 Nov;57(11):1140-8. doi: 10.1007/s11427-014-4752-9. Epub 2014 Oct 17. Sci China Life Sci. 2014. PMID: 25326069 Review. - Assessment of metagenomic assemblers based on hybrid reads of real and simulated metagenomic sequences.
Wang Z, Wang Y, Fuhrman JA, Sun F, Zhu S. Wang Z, et al. Brief Bioinform. 2020 May 21;21(3):777-790. doi: 10.1093/bib/bbz025. Brief Bioinform. 2020. PMID: 30860572 Free PMC article. Review.
Cited by
- Diversity of acid stress resistant variants of Listeria monocytogenes and the potential role of ribosomal protein S21 encoded by rpsU.
Metselaar KI, den Besten HM, Boekhorst J, van Hijum SA, Zwietering MH, Abee T. Metselaar KI, et al. Front Microbiol. 2015 May 8;6:422. doi: 10.3389/fmicb.2015.00422. eCollection 2015. Front Microbiol. 2015. PMID: 26005439 Free PMC article. - A Terpene Synthase Is Involved in the Synthesis of the Volatile Organic Compound Sodorifen of Serratia plymuthica 4Rx13.
Domik D, Thürmer A, Weise T, Brandt W, Daniel R, Piechulla B. Domik D, et al. Front Microbiol. 2016 May 19;7:737. doi: 10.3389/fmicb.2016.00737. eCollection 2016. Front Microbiol. 2016. PMID: 27242752 Free PMC article. - Maternal transmission, sex ratio distortion, and mitochondria.
Perlman SJ, Hodson CN, Hamilton PT, Opit GP, Gowen BE. Perlman SJ, et al. Proc Natl Acad Sci U S A. 2015 Aug 18;112(33):10162-8. doi: 10.1073/pnas.1421391112. Epub 2015 Apr 13. Proc Natl Acad Sci U S A. 2015. PMID: 25870270 Free PMC article. Review. - Identifying wrong assemblies in de novo short read primary sequence assembly contigs.
Chawla V, Kumar R, Shankar R. Chawla V, et al. J Biosci. 2016 Sep;41(3):455-74. doi: 10.1007/s12038-016-9630-0. J Biosci. 2016. PMID: 27581937 - Human milk metagenome: a functional capacity analysis.
Ward TL, Hosid S, Ioshikhes I, Altosaar I. Ward TL, et al. BMC Microbiol. 2013 May 25;13:116. doi: 10.1186/1471-2180-13-116. BMC Microbiol. 2013. PMID: 23705844 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources