MicroRNA functions in stress responses - PubMed (original) (raw)
Review
MicroRNA functions in stress responses
Anthony K L Leung et al. Mol Cell. 2010.
Abstract
MicroRNAs (miRNAs) are a class of ∼22 nucleotide short noncoding RNAs that play key roles in fundamental cellular processes, including how cells respond to changes in environment or, broadly defined, stresses. Responding to stresses, cells either choose to restore or reprogram their gene expression patterns. This decision is partly mediated by miRNA functions, in particular by modulating the amount of miRNAs, the amount of mRNA targets, or the activity/mode of action of miRNA-protein complexes. In turn, these changes determine the specificity, timing, and concentration of gene products expressed upon stresses. Dysregulation of these processes contributes to chronic diseases, including cancers.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures
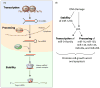
Figure 1. Biogenesis of mature miRNAs can be regulated at multiple levels
(A) miRNAs are first transcribed as primary transcripts (pri-miRNA), which fold into hairpin structures and are subsequently processed – first by Drosha in the nucleus and then by Dicer in the cytoplasm. These processing steps result in a ~22 nucleotide duplex in which one strand, the mature miRNA, binds to the core protein Argonaute and associated protein factors to form the final effector complex. The activity of the effector complexes is determined by the half-lives of the miRNAs, the protein composition of the effector complexes and the post-translational modifications of the individual components. TF: Transcription Factor; Pol II: RNA polymerase II. (B) Upon DNA damage, transcription, processing and stability of specific miRNAs are modulated, resulting in a p53-mediated gene expression program that promotes cell growth arrest and apoptosis.
Figure 2. Stress-induced expression of mRNA targets or target mimics affects miRNA activity
The protein output of an mRNA target is dependent on (A) the level of mRNA targets relative to the amount of miRNAs as well as (B) the level of mRNA targets relative to other mRNAs targeted by the same miRNA. (A) Translation of mRNA targets is generally suppressed by a constant level of miRNAs. However, when a particular mRNA target expression level exceeds the quantity of mRNA targets that can be titrated by the amount of miRNAs, translation of mRNA targets accelerates. (B) Translation of mRNA targets decreases along with the increase in miRNAs over time. Such decrease in translation halts as a result of the expression of target mimics, which compete with mRNA targets for the same miRNA.
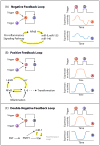
Figure 3. miRNAs are strategically located in (A) negative, (B) positive and (C) double-negative feedback loops to mediate stress responses
(A) A negative feedback loop: A (red) activates B (blue), but B represses A. This circuitry results in the restoration of the original level of A and B after the triggering stimulus is removed. For example, inflammation triggers a signaling cascade that results in a NF-κB-dependent transcription of a set of miRNAs. These miRNAs in turn target the components of the pro-inflammatory pathway, hence resetting the activation status of the inflammation pathway. (B) A positive feedback loop: in this circuit, A activates B and B activates A. Therefore, there could be a stable state with both A and B on or both A and B off. Upon inflammation, an increase in IL-6 triggers activation of NF-κB and expression of LIN-28, which results in the reduction of let-7 expression. As let-7 normally represses the expression of IL-6, this reduction in let-7 results in an increase in expression of IL-6, thus further propagating the cycle of events. Unlike a negative feedback loop, this circuitry exhibits a persistent, self-perpetuating response long after the triggering stimulus is removed. (C) A double-negative feedback loop: A represses B and B represses A. Thus, there could be a steady state with A on and B off, or vice versa, but not both on or off. This is found in the case of miR-7 and Yan in the differentiation of Drosophila eyes. Similar to a positive feedback loop, this results in a self-perpetuating response long after the triggering stimulus is removed. Examples of feedback loops are highlighted in yellow on the left side of the panel and hypothetical timings of the trigger and resultant expression/activity of protein A and B are indicated on the right. A feed-forward loop is circled by dotted lines in panel C. Panel designs were adapted from Ferrell (2002) Current Opinion in Cell Biology 14:140–8.
Figure 4. Timing of mRNA target expression can be modulated by the interplay amongst transcription factors, miRNAs and mRNA targets
Depending on whether a transcription factor (TF) activates or represses the level of miRNAs and/or mRNA targets, different timing of mRNA target expression results. The assumption is that it takes time for mature miRNAs to accumulate (deplete) upon transcriptional activation (repression) in suppressing (de-repressing) their mRNA targets. On the other hand, the change in the expression of mRNA targets is immediate.
Figure 5. Different ways to modulate miRNA activities upon stress
The miRNA function can be modulated at multiple levels by changing (A) the level of mature miRNAs, (B) the level of mRNA targets, (C) the activity of miRNA-protein complex and (D) the mode of action of miRNA-protein complex. (A) Shown is an mRNA target that has 3 binding sites for 3 different miRNAs. In normal condition, the target is repressed by miRNAs A and B. Upon stress, expression of mRNA target decreases if the level of miRNA C increases (hence 3 sites are bound by miRNAs); alternatively, the expression increases if the level of miRNA B decreases (only 1 site is bound). (B) The expression of mRNA targets (black) increases if the expression of mRNA target mimics (green) increases upon stress. These mimics compete with the same miRNAs as mRNA targets, thereby causing a relief in the repression of mRNA targets (“Target mimicry”). Alternatively, upon stress, cells could express different isoforms of the mRNA targets where miRNA binding sites could be created or deleted. (C) A change in the activity of miRNA-protein complex upon stress could be a result of post-translational modifications of components in the complex, direct association with other stress-specific co-factors, activation/repression from adjacent RNA-binding proteins bound on the same mRNA targets or sequestration to specific subcellular locales, such as stress granules or P-bodies. (D) Stress might alter the balance between the two major modes of action of miRNA-protein complexes: accelerating mRNA decay or inhibiting translation. Note that mRNA degradation is irreversible by nature and, hence, tipping towards this mode of action upon stress could potentially alter the composition of the transcriptome.
Similar articles
- Stress-induced changes in miRNA biogenesis and functioning.
Olejniczak M, Kotowska-Zimmer A, Krzyzosiak W. Olejniczak M, et al. Cell Mol Life Sci. 2018 Jan;75(2):177-191. doi: 10.1007/s00018-017-2591-0. Epub 2017 Jul 17. Cell Mol Life Sci. 2018. PMID: 28717872 Free PMC article. Review. - microRNA-1827 represses MDM2 to positively regulate tumor suppressor p53 and suppress tumorigenesis.
Zhang C, Liu J, Tan C, Yue X, Zhao Y, Peng J, Wang X, Laddha SV, Chan CS, Zheng S, Hu W, Feng Z. Zhang C, et al. Oncotarget. 2016 Feb 23;7(8):8783-96. doi: 10.18632/oncotarget.7088. Oncotarget. 2016. PMID: 26840028 Free PMC article. - Dynamics of microRNA biogenesis: crosstalk between p53 network and microRNA processing pathway.
Suzuki HI, Miyazono K. Suzuki HI, et al. J Mol Med (Berl). 2010 Nov;88(11):1085-94. doi: 10.1007/s00109-010-0650-1. Epub 2010 Jul 8. J Mol Med (Berl). 2010. PMID: 20614100 Review. - An integrative transcriptomic analysis reveals p53 regulated miRNA, mRNA, and lncRNA networks in nasopharyngeal carcinoma.
Gong Z, Yang Q, Zeng Z, Zhang W, Li X, Zu X, Deng H, Chen P, Liao Q, Xiang B, Zhou M, Li X, Li Y, Xiong W, Li G. Gong Z, et al. Tumour Biol. 2016 Mar;37(3):3683-95. doi: 10.1007/s13277-015-4156-x. Epub 2015 Oct 13. Tumour Biol. 2016. PMID: 26462838 - Loss of microRNA-143/145 disturbs cellular growth and apoptosis of human epithelial cancers by impairing the MDM2-p53 feedback loop.
Zhang J, Sun Q, Zhang Z, Ge S, Han ZG, Chen WT. Zhang J, et al. Oncogene. 2013 Jan 3;32(1):61-9. doi: 10.1038/onc.2012.28. Epub 2012 Feb 13. Oncogene. 2013. PMID: 22330136
Cited by
- MicroRNA mediation of endothelial inflammatory response to smooth muscle cells and its inhibition by atheroprotective shear stress.
Chen LJ, Chuang L, Huang YH, Zhou J, Lim SH, Lee CI, Lin WW, Lin TE, Wang WL, Chen L, Chien S, Chiu JJ. Chen LJ, et al. Circ Res. 2015 Mar 27;116(7):1157-69. doi: 10.1161/CIRCRESAHA.116.305987. Epub 2015 Jan 26. Circ Res. 2015. PMID: 25623956 Free PMC article. - Emerging delivery approaches for targeted pulmonary fibrosis treatment.
Diwan R, Bhatt HN, Beaven E, Nurunnabi M. Diwan R, et al. Adv Drug Deliv Rev. 2024 Jan;204:115147. doi: 10.1016/j.addr.2023.115147. Epub 2023 Dec 6. Adv Drug Deliv Rev. 2024. PMID: 38065244 Free PMC article. Review. - Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics.
Kim T, Xu Z, Clauder-Münster S, Steinmetz LM, Buratowski S. Kim T, et al. Cell. 2012 Sep 14;150(6):1158-69. doi: 10.1016/j.cell.2012.08.016. Epub 2012 Sep 6. Cell. 2012. PMID: 22959268 Free PMC article. - Characterization of grapevine microR164 and its target genes.
Sun X, Korir NK, Han J, Shangguan LF, Kayesh E, Leng XP, Fang JG. Sun X, et al. Mol Biol Rep. 2012 Oct;39(10):9463-72. doi: 10.1007/s11033-012-1811-9. Epub 2012 Jun 24. Mol Biol Rep. 2012. PMID: 22733489 - Feline microRNAome in ovary and testis: Exploration of in-silico miRNA-mRNA networks involved in gonadal function and cellular stress response.
Amelkina O, da Silva AM, Silva AR, Comizzoli P. Amelkina O, et al. Front Genet. 2022 Sep 26;13:1009220. doi: 10.3389/fgene.2022.1009220. eCollection 2022. Front Genet. 2022. PMID: 36226169 Free PMC article.
References
- Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. - PubMed
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-Mediated Translational Repression in Human Cells Subjected to Stress. Cell. 2006;125:1111–1124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01-CA133404/CA/NCI NIH HHS/United States
- P01 CA042063/CA/NCI NIH HHS/United States
- P30 CA014051/CA/NCI NIH HHS/United States
- P30-CA14051/CA/NCI NIH HHS/United States
- R01 CA133404/CA/NCI NIH HHS/United States
- P01-CA42063/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources