Fate mapping analysis reveals that adult microglia derive from primitive macrophages - PubMed (original) (raw)
. 2010 Nov 5;330(6005):841-5.
doi: 10.1126/science.1194637. Epub 2010 Oct 21.
Melanie Greter, Marylene Leboeuf, Sayan Nandi, Peter See, Solen Gokhan, Mark F Mehler, Simon J Conway, Lai Guan Ng, E Richard Stanley, Igor M Samokhvalov, Miriam Merad
Affiliations
- PMID: 20966214
- PMCID: PMC3719181
- DOI: 10.1126/science.1194637
Fate mapping analysis reveals that adult microglia derive from primitive macrophages
Florent Ginhoux et al. Science. 2010.
Abstract
Microglia are the resident macrophages of the central nervous system and are associated with the pathogenesis of many neurodegenerative and brain inflammatory diseases; however, the origin of adult microglia remains controversial. We show that postnatal hematopoietic progenitors do not significantly contribute to microglia homeostasis in the adult brain. In contrast to many macrophage populations, we show that microglia develop in mice that lack colony stimulating factor-1 (CSF-1) but are absent in CSF-1 receptor-deficient mice. In vivo lineage tracing studies established that adult microglia derive from primitive myeloid progenitors that arise before embryonic day 8. These results identify microglia as an ontogenically distinct population in the mononuclear phagocyte system and have implications for the use of embryonically derived microglial progenitors for the treatment of various brain disorders.
Figures
Fig. 1
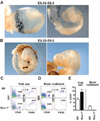
Microglia arise during early embryonic life. (A) Left image, schematic of the imaging field. Right image, three-dimensional rendering of E10.5 brain rudiment from Cx3cr1gfp/+ mice. DAPI (blue) stains the ectoderm. Representative data of two experiments. (B and C) Flow-cytometric analysis of the expression of CD11b and GFP (CX3CR1) on gated 4´,6´-diamidino-2-phenylindole (DAPI)–CD45+ brain (B) and yolk sac (C) cells isolated from Cx3cr1gfp/+ mice at different stages during development. Histograms show F4/80 (red) or isotype control (blue) on gated cells. Representative data of three experiments.
Fig. 2
Microglia and yolk sac macrophages are absent in _Csf-1r_−/− mice. (A) Flow-cytometric analysis of CSF-1R expression (red) on microglia and yolk sac macrophages (blue, isotype control). Representative data of three experiments. (B and C) Percentage of microglia (B) and yolk sac macrophages (C) in _Csf-1r_−/− (black squares) or control littermate (Wt) (white squares) FVB/NJ mice. Pooled data from three separate experiments. **P < 0.001; ***P < 0.0001. (D) Coronal sections of 3-week-old Wt, Csf-1op/op, and _Csf-1r_−/− brains of region boxed in the schematic stained for the microglial marker Iba1. DG indicates dentate gyrus; Cx, cerebral cortex; CA3, CA3 region of the hippocampus. Mean number of Iba1+ cells per field from three different brain regions is shown. Average of six fields (0.5 mm2) per region per genotype. Error bars represent mean ± SD of data from two pooled experiments. *P < 0.05; ****P < 0.00001.
Fig. 3
Microglia arise from primitive myeloid progenitors. Runx1Cre/wt:Rosa26R26R-eYFP mice were treated with 4′OHT to induce Cre-mediated recombination at E7.25 to E7.5 and analyzed at E10.5 [(A) to (C)] or at 8 weeks postbirth [(D) and (E)]. Controls are nontreated mice. (A) Flow-cytometric analysis from one representative embryo showing the percent recombination among yolk sac macrophages and microglial progenitors. (B and D) Pooled data from two experiments showing the percent recombination among yolk sac macrophages and microglial progenitors cells in embryos (B), and among monocytes and microglia in adult mice (n = 10) (D). (C) Correlation and regression analysis between the percent recombination in microglial progenitors and yolk sac macrophages. _r_2, coefficient of regression. (E) Percent recombination among monocytes, lung macrophages, and microglia in adult mice activated at different embryonic age. Error bars represent mean ± SEM of pooled data from two experiments (n = 8 to 16). Gating strategy for each leukocyte population is detailed in fig. S8.
Fig. 4
Runx1+ yolk sac progenitors seed the brain between E8.5 and E9.5 through blood circulation. (A and B) Runx1Cre/w:Rosa26R26R-LacZ embryos activated at E7.25 to E7.5 were isolated at E8.25 to E8.5 (A) or E9.25 to E9.5 (B) and processed for whole-mount LacZ staining as described in the materials and methods section. At E8.25 to E8.5, labeled cells are detected in the yolk sac but not in the brain rudiment or in the neural tube (A), whereas labeled cells infiltrate the brain rudiment of E9.25 to E9.5 embryos (B). (C to E) Yolk sac and brain rudiment tissues were isolated from E10.0 to E10.5 _Ncx1_−/− embryos or control littermates and processed for flow cytometry analysis as described in the materials and methods section. Dot plots show the presence of yolk sac macrophages in _Ncx1_−/− embryos and control littermates (C), whereas microglia were present in control but not in _Ncx1_−/− embryos (D). (E) The percentage ± SEM of hematopoietic cells (CD45+) in control littermates (white bars, n = 4) and _Ncx1_−/− embryos (black bars, n = 3).
Comment in
- Neuroimmunology: The origins of microglial cells.
Leavy O. Leavy O. Nat Rev Neurosci. 2010 Dec;11(12):787. doi: 10.1038/nrn2960. Nat Rev Neurosci. 2010. PMID: 21132880 No abstract available. - Microglial cell origins.
Leavy O. Leavy O. Nat Rev Immunol. 2010 Dec;10(12):808. doi: 10.1038/nri2896. Nat Rev Immunol. 2010. PMID: 21155194 No abstract available.
Similar articles
- [Microglia arise from extra-embryonic yolk sac primitive progenitors].
Ginhoux F, Merad M. Ginhoux F, et al. Med Sci (Paris). 2011 Aug-Sep;27(8-9):719-24. doi: 10.1051/medsci/2011278013. Epub 2011 Aug 31. Med Sci (Paris). 2011. PMID: 21880259 Review. French. - Microglial pilgrimage to the brain.
[No authors listed] [No authors listed] Nat Med. 2010 Dec;16(12):1380-1. doi: 10.1038/nm1210-1380. Nat Med. 2010. PMID: 21135848 No abstract available. - Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors.
Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, Garner H, Trouillet C, de Bruijn MF, Geissmann F, Rodewald HR. Gomez Perdiguero E, et al. Nature. 2015 Feb 26;518(7540):547-51. doi: 10.1038/nature13989. Epub 2014 Dec 3. Nature. 2015. PMID: 25470051 Free PMC article. - C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages.
Hoeffel G, Chen J, Lavin Y, Low D, Almeida FF, See P, Beaudin AE, Lum J, Low I, Forsberg EC, Poidinger M, Zolezzi F, Larbi A, Ng LG, Chan JK, Greter M, Becher B, Samokhvalov IM, Merad M, Ginhoux F. Hoeffel G, et al. Immunity. 2015 Apr 21;42(4):665-78. doi: 10.1016/j.immuni.2015.03.011. Immunity. 2015. PMID: 25902481 Free PMC article. - Fetal monocytes and the origins of tissue-resident macrophages.
Hoeffel G, Ginhoux F. Hoeffel G, et al. Cell Immunol. 2018 Aug;330:5-15. doi: 10.1016/j.cellimm.2018.01.001. Epub 2018 Jan 12. Cell Immunol. 2018. PMID: 29475558 Review.
Cited by
- Parenteral high‑dose ascorbate - A possible approach for the treatment of glioblastoma (Review).
Renner O, Burkard M, Michels H, Vollbracht C, Sinnberg T, Venturelli S. Renner O, et al. Int J Oncol. 2021 Jun;58(6):35. doi: 10.3892/ijo.2021.5215. Epub 2021 May 6. Int J Oncol. 2021. PMID: 33955499 Free PMC article. Review. - The Role of Microglia in Inherited White-Matter Disorders and Connections to Frontotemporal Dementia.
Sirkis DW, Bonham LW, Yokoyama JS. Sirkis DW, et al. Appl Clin Genet. 2021 Mar 31;14:195-207. doi: 10.2147/TACG.S245029. eCollection 2021. Appl Clin Genet. 2021. PMID: 33833548 Free PMC article. Review. - Immunological Markers for Central Nervous System Glia.
Huang H, He W, Tang T, Qiu M. Huang H, et al. Neurosci Bull. 2023 Mar;39(3):379-392. doi: 10.1007/s12264-022-00938-2. Epub 2022 Aug 26. Neurosci Bull. 2023. PMID: 36028641 Free PMC article. Review. - MMP-3 mediates psychosine-induced globoid cell formation: implications for leukodystrophy pathology.
Ijichi K, Brown GD, Moore CS, Lee JP, Winokur PN, Pagarigan R, Snyder EY, Bongarzone ER, Crocker SJ. Ijichi K, et al. Glia. 2013 May;61(5):765-77. doi: 10.1002/glia.22471. Epub 2013 Feb 13. Glia. 2013. PMID: 23404611 Free PMC article. - Brain-reactive antibodies and disease.
Diamond B, Honig G, Mader S, Brimberg L, Volpe BT. Diamond B, et al. Annu Rev Immunol. 2013;31:345-85. doi: 10.1146/annurev-immunol-020711-075041. Annu Rev Immunol. 2013. PMID: 23516983 Free PMC article. Review.
References
- Chan WY, Kohsaka S, Rezaie P. Brain Res. Brain Res. Rev. 2007;53:344. - PubMed
- Ransohoff RM, Perry VH. Annu. Rev. Immunol. 2009;27:119. - PubMed
- Materials and methods are available as supporting material on Science Online.
- Leong SK, Ling EA. Glia. 1992;6:39. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI080884/AI/NIAID NIH HHS/United States
- AI080884/AI/NIAID NIH HHS/United States
- R01 CA032551/CA/NCI NIH HHS/United States
- R37 CA026504/CA/NCI NIH HHS/United States
- CA112100/CA/NCI NIH HHS/United States
- R01 CA112100/CA/NCI NIH HHS/United States
- R01 HL060714/HL/NHLBI NIH HHS/United States
- R01 NS038902/NS/NINDS NIH HHS/United States
- NS38902/NS/NINDS NIH HHS/United States
- R01 HL086899/HL/NHLBI NIH HHS/United States
- R01 MH066290/MH/NIMH NIH HHS/United States
- HL086899/HL/NHLBI NIH HHS/United States
- CA32551/CA/NCI NIH HHS/United States
- CA26504/CA/NCI NIH HHS/United States
- R01 CA026504/CA/NCI NIH HHS/United States
- MH66290/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous