WebGeSTer DB--a transcription terminator database - PubMed (original) (raw)
. 2011 Jan;39(Database issue):D129-35.
doi: 10.1093/nar/gkq971. Epub 2010 Oct 23.
Affiliations
- PMID: 20972211
- PMCID: PMC3013805
- DOI: 10.1093/nar/gkq971
WebGeSTer DB--a transcription terminator database
Anirban Mitra et al. Nucleic Acids Res. 2011 Jan.
Abstract
We present WebGeSTer DB, the largest database of intrinsic transcription terminators (http://pallab.serc.iisc.ernet.in/gester). The database comprises of a million terminators identified in 1060 bacterial genome sequences and 798 plasmids. Users can obtain both graphic and tabular results on putative terminators based on default or user-defined parameters. The results are arranged in different tiers to facilitate retrieval, as per the specific requirements. An interactive map has been incorporated to visualize the distribution of terminators across the whole genome. Analysis of the results, both at the whole-genome level and with respect to terminators downstream of specific genes, offers insight into the prevalence of canonical and non-canonical terminators across different phyla. The data in the database reinforce the paradigm that intrinsic termination is a conserved and efficient regulatory mechanism in bacteria. Our database is freely accessible.
Figures
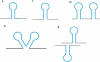
Figure 1.
Terminators catalogued in the WebGeSTer DB—(i) L-shaped (canonical terminators): hairpin + 10 bp trail having >3 uridylates, (ii) I-shaped: hairpin + ≤3 uridines in the trail, (iii) U-shaped: >1 hairpin in tandem with an interval of <50 nt in between, (iv) V-shaped: two hairpins’ structure, where the second structure starts immediately at the end of first one and (v) X-shaped: convergent type structures that function as terminators for the convergently transcribed genes on two different strands.
Figure 2.
GeSTerDB and WebGeSTer interface. The user can refine his search for terminator profiles with one or more of the criteria provided. For e.g. a search for ‘Total ORFs >5000’ and ‘Terminators (lowest Δ_G_)>2500’ would retrieve all genomes which have more than 5000 genes and also greater than 2500 ‘Best’ terminators.
Figure 3.
Progressive data accession in WebGeSTer DB. A search initiated at a specific genome can finally lead to details about a terminator downstream of a specific gene.
Figure 4.
TER-MAP—the high-resolution terminator map and browser. From the genome summary page, the user can navigate to a defined region of the genome or to a specific gene. Terminators are represented as ‘lollipops’ at ends of genes. Clicking on any terminator leads to more information about its computed structure and other parameters.
Similar articles
- Occurrence, divergence and evolution of intrinsic terminators across eubacteria.
Mitra A, Angamuthu K, Jayashree HV, Nagaraja V. Mitra A, et al. Genomics. 2009 Aug;94(2):110-6. doi: 10.1016/j.ygeno.2009.04.004. Epub 2009 Apr 23. Genomics. 2009. PMID: 19393739 - Genome-wide analysis of the intrinsic terminators of transcription across the genus Mycobacterium.
Mitra A, Angamuthu K, Nagaraja V. Mitra A, et al. Tuberculosis (Edinb). 2008 Nov;88(6):566-75. doi: 10.1016/j.tube.2008.06.004. Epub 2008 Sep 3. Tuberculosis (Edinb). 2008. PMID: 18768372 - Parameters affecting transcription termination by Escherichia coli RNA. II. Construction and analysis of hybrid terminators.
Reynolds R, Chamberlin MJ. Reynolds R, et al. J Mol Biol. 1992 Mar 5;224(1):53-63. doi: 10.1016/0022-2836(92)90575-5. J Mol Biol. 1992. PMID: 1372366 - Rho-dependent terminators and transcription termination.
Ciampi MS. Ciampi MS. Microbiology (Reading). 2006 Sep;152(Pt 9):2515-2528. doi: 10.1099/mic.0.28982-0. Microbiology (Reading). 2006. PMID: 16946247 Review. - Transcription attenuation in bacteria: theme and variations.
Naville M, Gautheret D. Naville M, et al. Brief Funct Genomics. 2010 Mar;9(2):178-89. doi: 10.1093/bfgp/elq008. Brief Funct Genomics. 2010. PMID: 20352660 Review.
Cited by
- Genome-scale analyses of Escherichia coli and Salmonella enterica AraC reveal noncanonical targets and an expanded core regulon.
Stringer AM, Currenti S, Bonocora RP, Baranowski C, Petrone BL, Palumbo MJ, Reilly AA, Zhang Z, Erill I, Wade JT. Stringer AM, et al. J Bacteriol. 2014 Feb;196(3):660-71. doi: 10.1128/JB.01007-13. Epub 2013 Nov 22. J Bacteriol. 2014. PMID: 24272778 Free PMC article. - Characterization of 582 natural and synthetic terminators and quantification of their design constraints.
Chen YJ, Liu P, Nielsen AA, Brophy JA, Clancy K, Peterson T, Voigt CA. Chen YJ, et al. Nat Methods. 2013 Jul;10(7):659-64. doi: 10.1038/nmeth.2515. Epub 2013 Jun 2. Nat Methods. 2013. PMID: 23727987 - Genomic Profiling of Non-O157 Shiga Toxigenic _Escherichia coli_-Infecting Bacteriophages from South Africa.
Bumunang EW, McAllister TA, Polo RO, Ateba CN, Stanford K, Schlechte J, Walker M, MacLean K, Niu YD. Bumunang EW, et al. Phage (New Rochelle). 2022 Dec 1;3(4):221-230. doi: 10.1089/phage.2022.0003. Epub 2022 Dec 19. Phage (New Rochelle). 2022. PMID: 36793886 Free PMC article. - Bacterial transcription terminators: the RNA 3'-end chronicles.
Peters JM, Vangeloff AD, Landick R. Peters JM, et al. J Mol Biol. 2011 Oct 7;412(5):793-813. doi: 10.1016/j.jmb.2011.03.036. Epub 2011 Mar 23. J Mol Biol. 2011. PMID: 21439297 Free PMC article. - Actinomycetes biosynthetic potential: how to bridge in silico and in vivo?
Rebets Y, Brötz E, Tokovenko B, Luzhetskyy A. Rebets Y, et al. J Ind Microbiol Biotechnol. 2014 Feb;41(2):387-402. doi: 10.1007/s10295-013-1352-9. Epub 2013 Oct 15. J Ind Microbiol Biotechnol. 2014. PMID: 24127068 Review.
References
- von Hippel PH, Delagoutte E. A general model for nucleic acid helicases and their “coupling” within macromolecular machines. Cell. 2001;104:177–190. - PubMed
- Platt T. Transcription termination and the regulation of gene expression. Annu. Rev. Biochem. 1986;55:339–372. - PubMed
- Borukhov S, Nudler E. RNA polymerase: the vehicle of transcription. Trends Microbiol. 2008;16:126–134. - PubMed
- Richardson JP, Greenblatt J. Control of RNA chain elongation and termination. In: Neidhert FC, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd Edn. Washington, DC: ASM press; 1996. pp. 822–848.
- Henkin TM. Control of transcription termination in prokaryotes. Annu. Rev. Genet. 1996;30:35–57. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous