SUMO E3 ligase activity of TRIM proteins - PubMed (original) (raw)
SUMO E3 ligase activity of TRIM proteins
Y Chu et al. Oncogene. 2011.
Abstract
SUMOylation governs numerous cellular processes and is essential to most eukaryotic life. Despite increasing recognition of the importance of this process, an extremely limited number of small ubiquitin-like modifier (SUMO) protein ligases (E3s) have been identified. Here we show that at least some members of the functionally diverse tripartite motif (TRIM) superfamily are SUMO E3s. These TRIM proteins bind both the SUMO-conjugating enzyme Ubc9 and substrates and strongly enhance transfer of SUMOs from Ubc9 to these substrates. Among the substrates of TRIM SUMO E3s are the tumor suppressor p53 and its principal antagonist Mdm2. The E3 activity depends on the TRIM motif, suggesting it to be the first widespread SUMO E3 motif. Given the large number of TRIM proteins, our results may greatly expand the identified SUMO E3s. Furthermore, TRIM E3 activity may be an important contributor to SUMOylation specificity and the versatile functions of TRIM proteins.
Conflict of interest statement
Conflict of interest
The authors declare no conflict of interest.
Figures
Figure 1. PML promotes SUMOylation of p53 and Mdm2 in vivo
(a) _Pml_−/− MEF cells were transfected with Flag-PML, p53, and SUMO1 as indicated. Cells were treated with proteasome inhibitor MG132 for 4 h (same below), and whole cell lysates (WCL) were analyzed by western blot. Molecular weight standards (in kDa) are shown on the left. (b) H1299 cells were transfected with Flag-PML, HA-p53, and His- or Flag-tagged SUMO1 as indicated. Cells were lysed in buffer containing guanidinium-HCl, and proteins conjugated to His-SUMO1 were pulled down by Ni2+-NTA beads (Ni beads). The bead-bound proteins and WCL were analyzed by western blot. (c) H1299 cells were transfected with PML isoform IV or VI, p53, SUMO1 and GFP as indicated. WCL were analyzed by western blot. (d) _Pml_−/− cells were transfected with HA-PML, Flag-Mdm2, and His-SUMO1 as indicated. Cell lysates were incubated by Ni2+-NTA beads. WCL (left) and the bead-bound proteins (right) were analyzed by western blot.
Figure 2. Purified PML promotes SUMOylation of p53 and Mdm2 in vitro
(a) His-p53 purified from bacteria was incubated with SUMO E1(SAE1/SAE2), ATP, and either SUMO1 or SUMO2, in the presence or absence of Ubc9 and PML. The reaction mixes were analyzed by western blot using anti-p53 antibody. (b) In vitro SUMOylation of Mdm2 was performed with SUMO E1, His-SUMO1, and ATP, in the presence or absence of Ubc9 and PML. Proteins conjugated to His-SUMO1 were purified by Ni-NTA beads and analyzed by western blot using anti-Mdm2 antibody. (c) His-p53 was incubated with ATP and the indicated combinations of E1, Ubc9, and bacterial recombinant GST-PML or GST. The reaction mixes were analyzed as in (a).
Figure 3. The Ring and B boxes domains are required for SUMO E3 activity of PML
(a) Schematic representation of wild type PML isoform IV (called PML in this study), isoform VI, and various PML mutants. The RING domain (R) (aa 59–91), B1 box (aa 124–166), B2 box (aa 185–230), predicted coiled-coil region (CC) (aa 230–323), and SIM (aa 508–511) are indicated. PML mutations are: N (aa 1–233), C (aa 223–633), Rm (Cys57-to-Ser and Cys60-to-Ser, C57A/C60A), B1m (C129A/C132A), B2m (C189A/H194A), M4 (combination of Rm and B1m), and M6 (combination of Rm, B1m, and B2m). Asterisks indicate the positions of the point mutations. (b) PML mutations affect Mdm2 SUMOylation. _Pml_−/− cells were transfected with PML or each of the PML mutants, Mdm2, and His-SUMO1 as indicated. His-SUMO1-conjugated proteins pulled down by Ni2+-NTA beads and WCL were analyzed by western blot. (c) _Pml_−/− MEF cells were transfected with p53, His-SUMO1, and Flag-tagged PML or each of the PML mutants. Cell lysates were analyzed by western blots. (d) In vitro SUMOylation of His-p53 (top) and Flag-Daxx (middle) by PML and PML point mutants. The relative levels of PML proteins are shown in the bottom. His-p53 was purified from bacteria and Flag-Daxx was purified from 293T cells.
Figure 4. Other TRIM proteins promote Mdm2 SUMOylation in vivo and in vitro
(a and b) 293T cells were transfected with His-SUMO1, HA-Mdm2, and EGFP unless indicated otherwise, together with the TRIM expression constructs or the corresponding vector controls (V). Proteins conjugated to His-SUMO1 and WCL were analyzed by western blot. (c) Summary of the effect of TRIM proteins on Mdm2 SUMOylation. The results are shown in (A) and (B), as well as in Figs. S3.
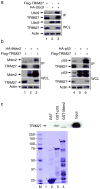
Figure 5. TRIM27 is associated with ubc9, p53 and Mdm2
(a) 293T cells were co-transfected with Flag-TRIM27 and/or HA-Ubc9. Cell lysates were incubated with anti-Flag M2 agarose. Immunoprecipitated proteins (IP) and whole cell lysates were analyzed by western blot. (b) 293T cells were co-transfected with Flag-TRIM27 and either HA-Mdm2 (left) or HA-p53 (right). Protein interactions were examined as in (a). *, a cleaved product of p53. (c) TRIM27 directly binds to p53. GST, GST-p53, and GST-Mdm2 immobilized on glutathione beads were incubated with Flag-TRIM27 purified from 293T cells. Top: The bound proteins and 10% of the input were analyzed by Western blotting with anti-Flag antibody. Bottom: GST proteins were analyzed by Coomassie staining. GST, full-length GST-p53 and GST-Mdm2 proteins are labeled by asterisks. The sizes of the molecular weight standards in the left lane are labeled (in kDa).
Figure 6. TRIM27 stabilizes Mdm2 through SUMOylation
(a) _Pml_−/− MEF cells were transfected with His-SUMO1, HA-Mdm2, TRIM27, PML or PIASy as indicated. His-SUMO1 conjugates and WCL were analyzed by western blot. (b) Bacterially expressed p53 was incubated with SUMO E1, His-SUMO1/His-SUMO2, in the presence or absence of Ubc9 and Flag-TRIM27. Reaction mixes were analyzed by western blot using anti-p53 antibody. (c) In vitro Mdm2 SUMOylation reactions were performed with SUMO E1, ATP, His-SUMO1/SUMO2, _in vitro_-translated Mdm2, in the presence or absence of Ubc9 and Flag-TRIM27 purified from 293T cells. Proteins conjugated to His-SUMO were captured by Ni2+-NTA beads pull down and analyzed by western blot with anti-Mdm2 antibody. (d) His-p53 was incubated with ATP and Ubc9, E1, and bacterial recombinant GST or GST-TRIM27 as indicated. The reaction mixes were analyzed by western blot using anti-p53 antibody. (e and f) HA-Mdm2 were transfected into HeLa cells together with Flag-TRIM27 and/or His-SUMO1. Cells were treated with cycloheximide for the indicated durations. (e) Cell lysates were analyzed for the levels of Mdm2. (f) The same samples were analyzed by western blot with different exposure times for each blot to achieve comparable band intensity at time 0. (g) Ubc9 down-regulation reduces Mdm2 level. U2OS cells treated with Ubc9 siRNA control or control siRNA were transfected with HA-Mdm2, Flag-TRIM27, His-SUMO1 as indicated.
Similar articles
- Kaposi's sarcoma-associated herpesvirus K-Rta exhibits SUMO-targeting ubiquitin ligase (STUbL) like activity and is essential for viral reactivation.
Izumiya Y, Kobayashi K, Kim KY, Pochampalli M, Izumiya C, Shevchenko B, Wang DH, Huerta SB, Martinez A, Campbell M, Kung HJ. Izumiya Y, et al. PLoS Pathog. 2013;9(8):e1003506. doi: 10.1371/journal.ppat.1003506. Epub 2013 Aug 22. PLoS Pathog. 2013. PMID: 23990779 Free PMC article. - Site-specific inhibition of the small ubiquitin-like modifier (SUMO)-conjugating enzyme Ubc9 selectively impairs SUMO chain formation.
Wiechmann S, Gärtner A, Kniss A, Stengl A, Behrends C, Rogov VV, Rodriguez MS, Dötsch V, Müller S, Ernst A. Wiechmann S, et al. J Biol Chem. 2017 Sep 15;292(37):15340-15351. doi: 10.1074/jbc.M117.794255. Epub 2017 Aug 7. J Biol Chem. 2017. PMID: 28784659 Free PMC article. - MEL-18 interacts with HSF2 and the SUMO E2 UBC9 to inhibit HSF2 sumoylation.
Zhang J, Goodson ML, Hong Y, Sarge KD. Zhang J, et al. J Biol Chem. 2008 Mar 21;283(12):7464-9. doi: 10.1074/jbc.M707122200. Epub 2008 Jan 21. J Biol Chem. 2008. PMID: 18211895 Free PMC article. - Molecular mechanisms in SUMO conjugation.
Varejão N, Lascorz J, Li Y, Reverter D. Varejão N, et al. Biochem Soc Trans. 2020 Feb 28;48(1):123-135. doi: 10.1042/BST20190357. Biochem Soc Trans. 2020. PMID: 31872228 Review. - Mechanisms and functions of SUMOylation in health and disease: a review focusing on immune cells.
Huang CH, Yang TT, Lin KI. Huang CH, et al. J Biomed Sci. 2024 Jan 27;31(1):16. doi: 10.1186/s12929-024-01003-y. J Biomed Sci. 2024. PMID: 38280996 Free PMC article. Review.
Cited by
- Crosstalk Between SUMO and Ubiquitin-Like Proteins: Implication for Antiviral Defense.
Chelbi-Alix MK, Thibault P. Chelbi-Alix MK, et al. Front Cell Dev Biol. 2021 Apr 21;9:671067. doi: 10.3389/fcell.2021.671067. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33968942 Free PMC article. Review. - SUMOylation of TUFT1 is essential for gastric cancer progression through AKT/mTOR signaling pathway activation.
Wang T, Min L, Gao Y, Zhao M, Feng S, Wang H, Wang Y, Zheng Y. Wang T, et al. Cancer Sci. 2023 Feb;114(2):533-545. doi: 10.1111/cas.15618. Epub 2022 Nov 15. Cancer Sci. 2023. PMID: 36380570 Free PMC article. - Paralogue-Specific Roles of SUMO1 and SUMO2/3 in Protein Quality Control and Associated Diseases.
Wang W, Matunis MJ. Wang W, et al. Cells. 2023 Dec 20;13(1):8. doi: 10.3390/cells13010008. Cells. 2023. PMID: 38201212 Free PMC article. Review. - Equol exerts anti-tumor effects on choriocarcinoma cells by promoting TRIM21-mediated ubiquitination of ANXA2.
Liu XM, Wang ZH, Wei QX, Song Y, Ma XX. Liu XM, et al. Biol Direct. 2024 Sep 6;19(1):78. doi: 10.1186/s13062-024-00519-5. Biol Direct. 2024. PMID: 39242533 Free PMC article. - TRIM27 negatively regulates NOD2 by ubiquitination and proteasomal degradation.
Zurek B, Schoultz I, Neerincx A, Napolitano LM, Birkner K, Bennek E, Sellge G, Lerm M, Meroni G, Söderholm JD, Kufer TA. Zurek B, et al. PLoS One. 2012;7(7):e41255. doi: 10.1371/journal.pone.0041255. Epub 2012 Jul 19. PLoS One. 2012. PMID: 22829933 Free PMC article.
References
- Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–1016. - PubMed
- Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat Cell Biol. 2004;6:665–672. - PubMed
- Cao T, Duprez E, Borden KL, Freemont PS, Etkin LD. Ret finger protein is a normal component of PML nuclear bodies and interacts directly with PML. J Cell Sci. 1998;111(Pt 10):1319–1329. - PubMed
- Chen L, Chen J. MDM2-ARF complex regulates p53 sumoylation. Oncogene. 2003;22:5348–5357. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous