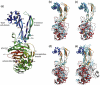Crystal structures of penicillin-binding protein 3 from Pseudomonas aeruginosa: comparison of native and antibiotic-bound forms - PubMed (original) (raw)
Comparative Study
Crystal structures of penicillin-binding protein 3 from Pseudomonas aeruginosa: comparison of native and antibiotic-bound forms
Sarah Sainsbury et al. J Mol Biol. 2011.
Abstract
We report the first crystal structures of a penicillin-binding protein (PBP), PBP3, from Pseudomonas aeruginosa in native form and covalently linked to two important β-lactam antibiotics, carbenicillin and ceftazidime. Overall, the structures of apo and acyl complexes are very similar; however, variations in the orientation of the amino-terminal membrane-proximal domain relative to that of the carboxy-terminal transpeptidase domain indicate interdomain flexibility. Binding of either carbenicillin or ceftazidime to purified PBP3 increases the thermostability of the enzyme significantly and is associated with local conformational changes, which lead to a narrowing of the substrate-binding cleft. The orientations of the two β-lactams in the active site and the key interactions formed between the ligands and PBP3 are similar despite differences in the two drugs, indicating a degree of flexibility in the binding site. The conserved binding mode of β-lactam-based inhibitors appears to extend to other PBPs, as suggested by a comparison of the PBP3/ceftazidime complex and the Escherichia coli PBP1b/ceftoxamine complex. Since P. aeruginosa is an important human pathogen, the structural data reveal the mode of action of the frontline antibiotic ceftazidime at the molecular level. Improved drugs to combat infections by P. aeruginosa and related Gram-negative bacteria are sought and our study provides templates to assist that process and allows us to discuss new ways of inhibiting PBPs.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures
Graphical abstract
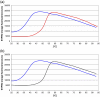
Fig. 1
Thermal shift assay. (a) PBP3 screened with 500 μM carbenicillin, giving a Δ_T_m of + 13.2 °C. The blue curve is the native protein, and the red curve is the complex. (b) PBP3 screened with 500 μM ceftazidime, giving a Δ_T_m of + 14.5 °C. The blue curve is the native protein, and the black curve is the complex.
Fig. 2
The overall structure of PBP3. (a) A ribbon diagram of the PBP3/carbenicillin complex showing the overall fold, antibiotic binding site, and bound glycerol. The diagram is rainbow-colored from blue at the N-terminus to red at the C-terminus, and the secondary structure elements are labelled in accordance with previous studies. (b–e) A comparison of the overall structure of native PBP3 (blue, orange, and red) with those of the PBP3/carbenicillin complex (b), PBP3/ceftazidime complex (c), PBP2 of N. gonorrhoeae (d), and PBP2x from S. pneumoniae (e). This figure was produced with PyMOL (
). The rest of the figures were drawn using BOBSCRIPT.
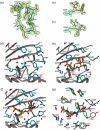
Fig. 3
The active site of PBP3. (a–c) 2_F_o − _F_c maps calculated after the final round of refinement and contoured at 1σ showing the electron density for the three active site structural motifs in apo-PBP3, bound carbenicillin, and ceftazidime in the two complexes, respectively. In (b) and (c), the carbon atoms of the acylated S294 are shown in cyan. (d–f) The active sites of apo-PBP3, carbenicillin, and ceftazidime complexes. The protein main chains are shown as gray ribbons, side chains are drawn as cyan sticks, and the inhibitor is shown in orange ball-and-sticks. The red spheres represent water molecules, and the yellow broken lines represent potential hydrogen bonds. Residue V333 that makes contacts with the bound inhibitors is not shown in (e) and (f) for clarity. (g) Comparison of the active sites of the PBP3/carbenicillin complex (cyan) and the PBP3/ceftazidime complex (orange).
Fig. 4
Glycerol binding, comparison with PBP1b, and electrostatic properties of the active site. (a) A diagram showing that the glycerol binding site is separated from the inhibitor binding site by β3–β4 strands and large conformational changes of the two strands due to inhibitor binding. The main-chain backbones of apo-PBP3 and the PBP3/carbenicillin complex are shown as gray and blue ribbons, respectively. (b) Comparison of the glycerol-binding site in the PBP3/carbenicillin complex with that in the apo-enzyme. The main chains and side chains are shown as ribbons and sticks, respectively (in gray and red for apo-PBP3; in blue and cyan for the carbenicillin complex), and the glycerol molecule is drawn as orange sticks. (c) Comparison of the active sites of the PBP3/ceftazidime complex (orange) and the S. pneumoniae PBP1b/cefotaxime complex (cyan). (d) Electrostatic surface showing the inhibitor-binding pocket of the PBP3/carbenicillin complex. (a) and (c) were produced with PyMOL (
).
Similar articles
- Structural Characterization of Diazabicyclooctane β-Lactam "Enhancers" in Complex with Penicillin-Binding Proteins PBP2 and PBP3 of Pseudomonas aeruginosa.
Rajavel M, Kumar V, Nguyen H, Wyatt J, Marshall SH, Papp-Wallace KM, Deshpande P, Bhavsar S, Yeole R, Bhagwat S, Patel M, Bonomo RA, van den Akker F. Rajavel M, et al. mBio. 2021 Feb 16;12(1):e03058-20. doi: 10.1128/mBio.03058-20. mBio. 2021. PMID: 33593978 Free PMC article. - Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Han S, Zaniewski RP, Marr ES, Lacey BM, Tomaras AP, Evdokimov A, Miller JR, Shanmugasundaram V. Han S, et al. Proc Natl Acad Sci U S A. 2010 Dec 21;107(51):22002-7. doi: 10.1073/pnas.1013092107. Epub 2010 Dec 6. Proc Natl Acad Sci U S A. 2010. PMID: 21135211 Free PMC article. - Structural analysis of the boronic acid β-lactamase inhibitor vaborbactam binding to Pseudomonas aeruginosa penicillin-binding protein 3.
Kumar V, Viviani SL, Ismail J, Agarwal S, Bonomo RA, van den Akker F. Kumar V, et al. PLoS One. 2021 Oct 15;16(10):e0258359. doi: 10.1371/journal.pone.0258359. eCollection 2021. PLoS One. 2021. PMID: 34653211 Free PMC article. - Development of new drugs for an old target: the penicillin binding proteins.
Zervosen A, Sauvage E, Frère JM, Charlier P, Luxen A. Zervosen A, et al. Molecules. 2012 Oct 24;17(11):12478-505. doi: 10.3390/molecules171112478. Molecules. 2012. PMID: 23095893 Free PMC article. Review.
Cited by
- Structural basis of Pseudomonas aeruginosa penicillin binding protein 3 inhibition by the siderophore-antibiotic cefiderocol.
Smith HG, Basak S, Aniebok V, Beech MJ, Alshref FM, Allen MD, Farley AJM, Schofield CJ. Smith HG, et al. Chem Sci. 2024 Sep 18;15(41):16928-37. doi: 10.1039/d4sc04937c. Online ahead of print. Chem Sci. 2024. PMID: 39328188 Free PMC article. - High-Throughput Crystallography Reveals Boron-Containing Inhibitors of a Penicillin-Binding Protein with Di- and Tricovalent Binding Modes.
Newman H, Krajnc A, Bellini D, Eyermann CJ, Boyle GA, Paterson NG, McAuley KE, Lesniak R, Gangar M, von Delft F, Brem J, Chibale K, Schofield CJ, Dowson CG. Newman H, et al. J Med Chem. 2021 Aug 12;64(15):11379-11394. doi: 10.1021/acs.jmedchem.1c00717. Epub 2021 Jul 31. J Med Chem. 2021. PMID: 34337941 Free PMC article. - Glycosyltransferases and Transpeptidases/Penicillin-Binding Proteins: Valuable Targets for New Antibacterials.
Sauvage E, Terrak M. Sauvage E, et al. Antibiotics (Basel). 2016 Feb 17;5(1):12. doi: 10.3390/antibiotics5010012. Antibiotics (Basel). 2016. PMID: 27025527 Free PMC article. Review. - Synthesis and biochemical evaluation of cephalosporin analogues equipped with chemical tethers.
Miller LM, Herman R, Gyulev I, Krauss TF, Thomas GH, Duhme-Klair AK. Miller LM, et al. RSC Adv. 2020 Oct 2;10(60):36485-36494. doi: 10.1039/d0ra04893c. eCollection 2020 Oct 1. RSC Adv. 2020. PMID: 35517937 Free PMC article. - Structural and biochemical analysis of the metallo-β-lactamase L1 from emerging pathogen Stenotrophomonas maltophilia revealed the subtle but distinct di-metal scaffold for catalytic activity.
Kim Y, Maltseva N, Wilamowski M, Tesar C, Endres M, Joachimiak A. Kim Y, et al. Protein Sci. 2020 Mar;29(3):723-743. doi: 10.1002/pro.3804. Epub 2019 Dec 24. Protein Sci. 2020. PMID: 31846104 Free PMC article.
References
- Sauvage E., Kerff F., Terrak M., Ayala J.A., Charlier P. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008;32:234–258. - PubMed
- Zapun A., Contreras-Martel C., Vernet T. Penicillin-binding proteins and beta-lactam resistance. FEMS Microbiol. Rev. 2008;32:361–385. - PubMed
- Powell A.J., Tomberg J., Deacon A.M., Nicholas R.A., Davies C. Crystal structures of penicillin-binding protein 2 from penicillin-susceptible and -resistant strains of Neisseria gonorrhoeae reveal an unexpectedly subtle mechanism for antibiotic resistance. J. Biol. Chem. 2009;284:1202–1212. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 083481/WT_/Wellcome Trust/United Kingdom
- BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G1100525/MRC_/Medical Research Council/United Kingdom
- G19/3/MRC_/Medical Research Council/United Kingdom
- G1000099/MRC_/Medical Research Council/United Kingdom
- 082596/WT_/Wellcome Trust/United Kingdom
- 075491/Z/04/WT_/Wellcome Trust/United Kingdom
- G0701506/MRC_/Medical Research Council/United Kingdom
- G0500365/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous