Epigenetic choreographers of neurogenesis in the adult mammalian brain - PubMed (original) (raw)
Review
Epigenetic choreographers of neurogenesis in the adult mammalian brain
Dengke K Ma et al. Nat Neurosci. 2010 Nov.
Abstract
Epigenetic mechanisms regulate cell differentiation during embryonic development and also serve as important interfaces between genes and the environment in adulthood. Neurogenesis in adults, which generates functional neural cell types from adult neural stem cells, is dynamically regulated by both intrinsic state-specific cell differentiation cues and extrinsic neural niche signals. Epigenetic regulation by DNA and histone modifiers, non-coding RNAs and other self-sustained mechanisms can lead to relatively long-lasting biological effects and maintain functional neurogenesis throughout life in discrete regions of the mammalian brain. Here, we review recent evidence that epigenetic mechanisms carry out diverse roles in regulating specific aspects of adult neurogenesis and highlight the implications of such epigenetic regulation for neural plasticity and disorders.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Figures
Figure 1
Basic modes of epigenetic regulation implicated in adult neurogenesis. (a) To initiate epigenetic processes, extracellular and intracellular signals may trigger epigenetic ‘perpetuators’ that form self-sustaining feedback loops or intrinsically produce long-lasting cellular effects in the absence of the initial trigger stimuli. Typical mechanisms by which this process occurs include transcription regulator and non-coding RNA–mediated feedback pathways, DNA methylation with associated methyl-binding proteins (MBDs), and histone H3K27 methylation with associated PcG (polycomb group) and TrxG (trithorax group) complexes. (b) DNA modifications. DNA methyltransferases (DNMTs) catalyze DNA methylation, whereas the pathway leading to DNA demethylation might include 5-methylcytosine (5mC) hydroxylase TET (ten-eleven translocation-1) proteins and DNA excision repair enzymes that are regulated by Gadd45 (growth arrest and DNA-damage-inducible) family proteins. (c) Histone modifications. Specific amino acid residues of histone N-terminal tails can be reversibly modified with a variety of ‘tags’ including acetylation (ac), phosphorylation (p), methylation (me), ubiquitination (ub), SUMOylation (su) and isomerization (iso). The varying turnover rates and biological interpreters of these modifications might execute different cellular functions for epigenetic regulation. C, cytosine; 5mC, 5-methylcytosines; 5hmC, 5-hydroxymethylcytosine; BER, base-excision repair; NER, nucleotide-excision repair; K, lysine; S, serine; T, threonine; R, arginine; P, proline; KAT, lysine acetyltransferase; HDAC, histone deacetylase; KMT, lysine methyltransferase; KDM, lysine deacetylase; PRMT, protein arginine methyltransferase; PADI4, peptidyl arginine deiminase type IV; JMJD6, Fe(II) and 2-oxoglutarate–dependent dioxygenase Jumonji domain-6 protein; DUB, deubiquitinase; SENP, sentrin-specific protease.
Figure 2
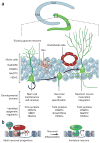
Classic and emerging technologies for epigenetic analysis of adult neurogenesis. During adult neurogenesis in the SGZ, neural stem cells (nestin+ and GFAP+) differentiate into immediate neural progenitors (Trb2+) and then newborn neurons (DCX+ and PSA-NCAM+) and finally into mature new neurons (NeuN+). To profile gene-specific epigenetic modifications, homogeneous target cell populations should be isolated and purified using laser capture microdissection or various prospective cell-labeling strategies. DNA methylation analysis can then be performed using bisulfite sequencing or methylation-sensitive restriction-based approaches. ChIP can be used to profile both DNA and histone modifications. The emerging technology of next-generation sequencing platforms allows rapid, genome-scale, high-resolution mapping of both DNA and histone modifications. RNA expression profiling may validate and further reveal new biological signatures for activated or repressed chromatin states.
Figure 3
Major epigenetic regulators of adult neurogenesis. (a) Current understanding of epigenetic regulation of adult neurogenesis in SGZ and SVZ. Adult neural progenitors undergo proliferation and generate neuroblasts that can further differentiate into mature, functional neurons. Neurogenesis can be regulated by intrinsic epigenetic mechanisms within the neuronal lineage of adult neural progenitors and extrinsically by nearby niche signaling cells, such as mature neurons, astrocytes and endothelial cells. During the stages of adult neurogenesis, diverse epigenetic mechanisms use common or different sets of molecules, including DNMT, PcG and TrxG, HDAC, MBD and Gadd45 family proteins, in either adult neural progenitors and their progeny or nearby niche cells to choreograph cell state transitions in coordination with other internal and external cues. Common molecules, such as HDACs, are likely to have different partners or binding sites depending on the differentiation stage during neurogenesis. Prototypical cell stages are shown to reflect identified epigenetic regulators for both SVZ and SGZ adult neurogenesis. (b) During early stages of adult hippocampal neurogenesis in the SGZ, L1 transcription is regulated by the transcription factors Sox2 and TCF to maintain the long-term silencing state, even during cell division. In response to external stimuli such as neuronal activity or Wnt signaling, the balance of co-repressor and co-activator in the L1 promoter is tipped to activation of L1 expression, driving its genomic retrotransposition. After terminal differentiation, adult neural progenitor–specific expression of Sox2 is downregulated and L1 transcription and retrotransposition are decreased. DNA methylation and MBD-mediated epigenetic mechanisms could act to ensure the long-term silencing of L1 by recruiting transcriptional co-repressors. CoA, co-activator; CoR, co-repressor; DG, dentate gyrus.
Similar articles
- Epigenetic regulation of neurogenesis in the adult mammalian brain.
Sun J, Sun J, Ming GL, Song H. Sun J, et al. Eur J Neurosci. 2011 Mar;33(6):1087-93. doi: 10.1111/j.1460-9568.2011.07607.x. Eur J Neurosci. 2011. PMID: 21395852 Free PMC article. Review. - Epigenetics, hippocampal neurogenesis, and neuropsychiatric disorders: unraveling the genome to understand the mind.
Hsieh J, Eisch AJ. Hsieh J, et al. Neurobiol Dis. 2010 Jul;39(1):73-84. doi: 10.1016/j.nbd.2010.01.008. Epub 2010 Jan 28. Neurobiol Dis. 2010. PMID: 20114075 Free PMC article. Review. - Genetics and Epigenetics in Adult Neurogenesis.
Hsieh J, Zhao X. Hsieh J, et al. Cold Spring Harb Perspect Biol. 2016 Jun 1;8(6):a018911. doi: 10.1101/cshperspect.a018911. Cold Spring Harb Perspect Biol. 2016. PMID: 27143699 Free PMC article. Review. - Epigenetic mechanisms in neurogenesis.
Yao B, Christian KM, He C, Jin P, Ming GL, Song H. Yao B, et al. Nat Rev Neurosci. 2016 Sep;17(9):537-49. doi: 10.1038/nrn.2016.70. Epub 2016 Jun 23. Nat Rev Neurosci. 2016. PMID: 27334043 Free PMC article. Review. - Adult neurogenesis and neurodegenerative diseases: A systems biology perspective.
Horgusluoglu E, Nudelman K, Nho K, Saykin AJ. Horgusluoglu E, et al. Am J Med Genet B Neuropsychiatr Genet. 2017 Jan;174(1):93-112. doi: 10.1002/ajmg.b.32429. Epub 2016 Feb 16. Am J Med Genet B Neuropsychiatr Genet. 2017. PMID: 26879907 Free PMC article.
Cited by
- Tet1 regulates adult hippocampal neurogenesis and cognition.
Zhang RR, Cui QY, Murai K, Lim YC, Smith ZD, Jin S, Ye P, Rosa L, Lee YK, Wu HP, Liu W, Xu ZM, Yang L, Ding YQ, Tang F, Meissner A, Ding C, Shi Y, Xu GL. Zhang RR, et al. Cell Stem Cell. 2013 Aug 1;13(2):237-45. doi: 10.1016/j.stem.2013.05.006. Epub 2013 Jun 13. Cell Stem Cell. 2013. PMID: 23770080 Free PMC article. - Preserved neurogenesis in non-demented individuals with AD neuropathology.
Briley D, Ghirardi V, Woltjer R, Renck A, Zolochevska O, Taglialatela G, Micci MA. Briley D, et al. Sci Rep. 2016 Jun 14;6:27812. doi: 10.1038/srep27812. Sci Rep. 2016. PMID: 27298190 Free PMC article. - Regulation of Adult Neurogenesis by Non-coding RNAs: Implications for Substance Use Disorders.
Oliver RJ, Mandyam CD. Oliver RJ, et al. Front Neurosci. 2018 Nov 22;12:849. doi: 10.3389/fnins.2018.00849. eCollection 2018. Front Neurosci. 2018. PMID: 30524229 Free PMC article. Review. - Global Exploration of RNA-Binding Proteins in Exercise-Induced Adult Hippocampal Neurogenesis: A Transcriptome Meta-analysis and Computational Study.
Nishanth MJ, Jha S. Nishanth MJ, et al. Biochem Genet. 2022 Dec;60(6):2471-2488. doi: 10.1007/s10528-022-10230-7. Epub 2022 May 11. Biochem Genet. 2022. PMID: 35546218 - Neuronal activity modifies the DNA methylation landscape in the adult brain.
Guo JU, Ma DK, Mo H, Ball MP, Jang MH, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, Zhang K, Ming GL, Gao Y, Song H. Guo JU, et al. Nat Neurosci. 2011 Aug 28;14(10):1345-51. doi: 10.1038/nn.2900. Nat Neurosci. 2011. PMID: 21874013 Free PMC article.
References
- Cameron HA, McKay RD. Adult neurogenesis produces a large pool of new granule cells in the dentate gyrus. J Comp Neurol. 2001;435:406–417. - PubMed
- Altman J, Das GD. Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J Comp Neurol. 1965;124:319–335. - PubMed
- Gage FH. Mammalian neural stem cells. Science. 2000;287:1433–1438. - PubMed
- Alvarez-Buylla A, Lim DA. For the long run: maintaining germinal niches in the adult brain. Neuron. 2004;41:683–686. - PubMed
- Ming GL, Song H. Adult neurogenesis in the mammalian central nervous system. Annu Rev Neurosci. 2005;28:223–250. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 HD069184/HD/NICHD NIH HHS/United States
- R01 MH090258-01/MH/NIMH NIH HHS/United States
- R01 AG024984/AG/NIA NIH HHS/United States
- MH090258/MH/NIMH NIH HHS/United States
- R37 NS047344/NS/NINDS NIH HHS/United States
- MH087874/MH/NIMH NIH HHS/United States
- R56 NS047344/NS/NINDS NIH HHS/United States
- R01 NS048271/NS/NINDS NIH HHS/United States
- R01 NS047344/NS/NINDS NIH HHS/United States
- R01 MH088485/MH/NIMH NIH HHS/United States
- R01 MH090258/MH/NIMH NIH HHS/United States
- R21 MH087874/MH/NIMH NIH HHS/United States
- R01 MH088485-02/MH/NIMH NIH HHS/United States
- AG024984/AG/NIA NIH HHS/United States
- R33 MH087874/MH/NIMH NIH HHS/United States
- NS048271/NS/NINDS NIH HHS/United States
- MH088485/MH/NIMH NIH HHS/United States
- HD069184/HD/NICHD NIH HHS/United States
- NS047344/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources