The phage-host arms race: shaping the evolution of microbes - PubMed (original) (raw)
Review
The phage-host arms race: shaping the evolution of microbes
Adi Stern et al. Bioessays. 2011 Jan.
Abstract
Bacteria, the most abundant organisms on the planet, are outnumbered by a factor of 10 to 1 by phages that infect them. Faced with the rapid evolution and turnover of phage particles, bacteria have evolved various mechanisms to evade phage infection and killing, leading to an evolutionary arms race. The extensive co-evolution of both phage and host has resulted in considerable diversity on the part of both bacterial and phage defensive and offensive strategies. Here, we discuss the unique and common features of phage resistance mechanisms and their role in global biodiversity. The commonalities between defense mechanisms suggest avenues for the discovery of novel forms of these mechanisms based on their evolutionary traits.
Figures
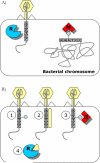
Figure 1. The restriction-modification defense system
A: A general illustration of function, exemplified by type II R–M enzymes. B: Examples of strategies employed by phage to evade restriction. (1) Incorporation of unusual bases protects from restriction [15]; (2) Masking of the restriction sites by phage proteins[112]; (3) Stimulation of MTase activity causes the phage DNA to be protected; (4) Neutralization of REase by phage proteins that mimic DNA [113].
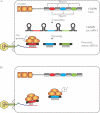
Figure 2. The CRISPR/Cas system
A: Mechanism of action: transcription from the repeat-spacer CRISPR locus generates a long non-coding RNA, with repeats that may sometimes assume a secondary structure. Cleavage of the repeat sequences by the Cas proteins generates crRNAs that target the phage DNA or RNA, and interfere with phage infection. B: Phages can evade CRISPR interference by mutation or recombination of the targeted proto-spacer sequence. Another putative evasion mechanism is phosphorylation of the Cas proteins. This remains, however, to be verified.
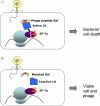
Figure 3. The abortive infection system
A: Illustration of the mechanism of the E. coli K-12 abortive infection Lit system. When the T4 phage peptide Gol is synthesized, it binds and activates the bacterial (prophage-encoded) Lit protein, which then cleaves the elongation factor EF-Tu. This leads to the arrest of protein synthesis and to bacterial cell death, with the phage trapped inside. B: A mutation at the Gol polypeptide reduces the activation of Lit, and rescues the phage.
Similar articles
- Phage Life Cycles Behind Bacterial Biodiversity.
Olszak T, Latka A, Roszniowski B, Valvano MA, Drulis-Kawa Z. Olszak T, et al. Curr Med Chem. 2017 Nov 24;24(36):3987-4001. doi: 10.2174/0929867324666170413100136. Curr Med Chem. 2017. PMID: 28412903 Review. - The Significance of Mutualistic Phages for Bacterial Ecology and Evolution.
Obeng N, Pratama AA, Elsas JDV. Obeng N, et al. Trends Microbiol. 2016 Jun;24(6):440-449. doi: 10.1016/j.tim.2015.12.009. Epub 2016 Jan 27. Trends Microbiol. 2016. PMID: 26826796 Review. - Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities.
Koskella B, Brockhurst MA. Koskella B, et al. FEMS Microbiol Rev. 2014 Sep;38(5):916-31. doi: 10.1111/1574-6976.12072. Epub 2014 Mar 27. FEMS Microbiol Rev. 2014. PMID: 24617569 Free PMC article. Review. - Bacteria-virus coevolution.
Buckling A, Brockhurst M. Buckling A, et al. Adv Exp Med Biol. 2012;751:347-70. doi: 10.1007/978-1-4614-3567-9_16. Adv Exp Med Biol. 2012. PMID: 22821466 Review. - Statistical structure of host-phage interactions.
Flores CO, Meyer JR, Valverde S, Farr L, Weitz JS. Flores CO, et al. Proc Natl Acad Sci U S A. 2011 Jul 12;108(28):E288-97. doi: 10.1073/pnas.1101595108. Epub 2011 Jun 27. Proc Natl Acad Sci U S A. 2011. PMID: 21709225 Free PMC article.
Cited by
- A Primary Physiological Role of Toxin/Antitoxin Systems Is Phage Inhibition.
Song S, Wood TK. Song S, et al. Front Microbiol. 2020 Aug 13;11:1895. doi: 10.3389/fmicb.2020.01895. eCollection 2020. Front Microbiol. 2020. PMID: 32903830 Free PMC article. Review. - Structure and rational engineering of the PglX methyltransferase and specificity factor for BREX phage defence.
Went SC, Picton DM, Morgan RD, Nelson A, Brady A, Mariano G, Dryden DTF, Smith DL, Wenner N, Hinton JCD, Blower TR. Went SC, et al. Nat Commun. 2024 Aug 22;15(1):7236. doi: 10.1038/s41467-024-51629-7. Nat Commun. 2024. PMID: 39174540 Free PMC article. - Functional Diversity of Cytotoxic tRNase/Immunity Protein Complexes from Burkholderia pseudomallei.
Johnson PM, Gucinski GC, Garza-Sánchez F, Wong T, Hung LW, Hayes CS, Goulding CW. Johnson PM, et al. J Biol Chem. 2016 Sep 9;291(37):19387-400. doi: 10.1074/jbc.M116.736074. Epub 2016 Jul 20. J Biol Chem. 2016. PMID: 27445337 Free PMC article. - A virocentric perspective on the evolution of life.
Koonin EV, Dolja VV. Koonin EV, et al. Curr Opin Virol. 2013 Oct;3(5):546-57. doi: 10.1016/j.coviro.2013.06.008. Epub 2013 Jul 12. Curr Opin Virol. 2013. PMID: 23850169 Free PMC article. Review. - Sequence, structure and functional diversity of PD-(D/E)XK phosphodiesterase superfamily.
Steczkiewicz K, Muszewska A, Knizewski L, Rychlewski L, Ginalski K. Steczkiewicz K, et al. Nucleic Acids Res. 2012 Aug;40(15):7016-45. doi: 10.1093/nar/gks382. Epub 2012 May 25. Nucleic Acids Res. 2012. PMID: 22638584 Free PMC article.
References
- Riesenfeld CS, Schloss PD, Handelsman J. Metagenomics: genomic analysis of microbial communities. Annu Rev Genet. 2004;38:525–52. - PubMed
- Bergh O, Borsheim KY, Bratbak G, Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI082376/AI/NIAID NIH HHS/United States
- R01 AI082376-01/AI/NIAID NIH HHS/United States
- R01 AI082376-04/AI/NIAID NIH HHS/United States
- R01AI082376-01/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases