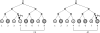SNP detection and genotyping from low-coverage sequencing data on multiple diploid samples - PubMed (original) (raw)
SNP detection and genotyping from low-coverage sequencing data on multiple diploid samples
Si Quang Le et al. Genome Res. 2011 Jun.
Abstract
Reductions in the cost of sequencing have enabled whole-genome sequencing to identify sequence variants segregating in a population. An efficient approach is to sequence many samples at low coverage, then to combine data across samples to detect shared variants. Here, we present methods to discover and genotype single-nucleotide polymorphism (SNP) sites from low-coverage sequencing data, making use of shared haplotype (linkage disequilibrium) information. For each population, we first collect SNP candidates based on independent sequence calls per site. We then use MARGARITA with genotype or phased haplotype data from the same samples to collect 20 ancestral recombination graphs (ARGs). We refine the posterior probability of SNP candidates by considering possible mutations at internal branches of the 40 marginal ancestral trees inferred from the 20 ARGs at the left and right flanking genotype sites. Using a population genetic prior distribution on tree-branch length and Bayesian inference, we determine a posterior probability of the SNP being real and also the most probable phased genotype call for each individual. We present experiments on both simulation data and real data from the 1000 Genomes Project to prove the applicability of the methods. We also explore the relative tradeoff between sequencing depth and the number of sequenced samples.
Figures
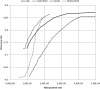
Figure 1.
Discovery and false-positive rates of QCALL for 400 samples with 4.0× coverage sequencing data. LDA and LDA,FW10 stand for linkage disequilibrium analysis without FW10 and with FW10. The same notation is applied to NLDA.
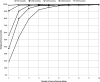
Figure 2.
SNP discovery power for different sequencing strategies, all using 1600× data, plotted as a function of the number of non-reference alleles present in the sequenced samples.
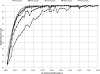
Figure 3.
SNP discovery power for different sequencing strategies as a function of the non-reference allele frequency in the population. The continuous lines show empirical results from the simulation with the allele frequency estimated from all 3000 simulated haplotypes, and the dashed lines present calculations based on sampling with marginal discovery rates per sample from Figure 2.
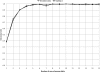
Figure 4.
Marginal discovery rates as a function of non-reference allele count in 43 samples, from the CEU simulation and from 1000 Genomes Project data evaluated at HapMap 2 sites not in HapMap 3, on the 43 sequenced samples overlapping HapMap 2.
Figure 5.
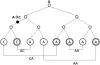
An illustrative example of a coalescent tree for four samples (eight haplotypes). Given a value at the root, A in this example, and a mutation from A to C in this example, we can infer genotypes for the four samples and, hence, compute the probability of data D conditional on this configuration. We estimate the likelihood of D given a tree t, p(D | t), by summing over all possible root values and mutations in t.
Figure 6.
Two mutations at two edges of a singleton (edges connected to haplotypes fourth or eighth) lead to the same genotype configuration.
Similar articles
- Linkage disequilibrium based genotype calling from low-coverage shotgun sequencing reads.
Duitama J, Kennedy J, Dinakar S, Hernández Y, Wu Y, Măndoiu II. Duitama J, et al. BMC Bioinformatics. 2011 Feb 15;12 Suppl 1(Suppl 1):S53. doi: 10.1186/1471-2105-12-S1-S53. BMC Bioinformatics. 2011. PMID: 21342586 Free PMC article. - A dynamic Bayesian Markov model for phasing and characterizing haplotypes in next-generation sequencing.
Zhang Y. Zhang Y. Bioinformatics. 2013 Apr 1;29(7):878-85. doi: 10.1093/bioinformatics/btt065. Epub 2013 Feb 13. Bioinformatics. 2013. PMID: 23407359 Free PMC article. - Linked region detection using high-density SNP genotype data via the minimum recombinant model of pedigree haplotype inference.
Wang L, Wang Z, Yang W. Wang L, et al. BMC Bioinformatics. 2009 Jul 15;10:216. doi: 10.1186/1471-2105-10-216. BMC Bioinformatics. 2009. PMID: 19604391 Free PMC article. - Genotype and SNP calling from next-generation sequencing data.
Nielsen R, Paul JS, Albrechtsen A, Song YS. Nielsen R, et al. Nat Rev Genet. 2011 Jun;12(6):443-51. doi: 10.1038/nrg2986. Nat Rev Genet. 2011. PMID: 21587300 Free PMC article. Review. - A review of software for microarray genotyping.
Lamy P, Grove J, Wiuf C. Lamy P, et al. Hum Genomics. 2011 May;5(4):304-9. doi: 10.1186/1479-7364-5-4-304. Hum Genomics. 2011. PMID: 21712191 Free PMC article. Review.
Cited by
- Evaluation of Whole-Exome Enrichment Solutions: Lessons from the High-End of the Short-Read Sequencing Scale.
Díaz-de Usera A, Lorenzo-Salazar JM, Rubio-Rodríguez LA, Muñoz-Barrera A, Guillen-Guio B, Marcelino-Rodríguez I, García-Olivares V, Mendoza-Alvarez A, Corrales A, Íñigo-Campos A, González-Montelongo R, Flores C. Díaz-de Usera A, et al. J Clin Med. 2020 Nov 13;9(11):3656. doi: 10.3390/jcm9113656. J Clin Med. 2020. PMID: 33202991 Free PMC article. - Next-generation sequencing of experimental mouse strains.
Yalcin B, Adams DJ, Flint J, Keane TM. Yalcin B, et al. Mamm Genome. 2012 Oct;23(9-10):490-8. doi: 10.1007/s00335-012-9402-6. Epub 2012 Jul 7. Mamm Genome. 2012. PMID: 22772437 Free PMC article. Review. - cn.MOPS: mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate.
Klambauer G, Schwarzbauer K, Mayr A, Clevert DA, Mitterecker A, Bodenhofer U, Hochreiter S. Klambauer G, et al. Nucleic Acids Res. 2012 May;40(9):e69. doi: 10.1093/nar/gks003. Epub 2012 Feb 1. Nucleic Acids Res. 2012. PMID: 22302147 Free PMC article. - A method for allocating low-coverage sequencing resources by targeting haplotypes rather than individuals.
Ros-Freixedes R, Gonen S, Gorjanc G, Hickey JM. Ros-Freixedes R, et al. Genet Sel Evol. 2017 Oct 25;49(1):78. doi: 10.1186/s12711-017-0353-y. Genet Sel Evol. 2017. PMID: 29070022 Free PMC article. - Efficient phasing and imputation of low-coverage sequencing data using large reference panels.
Rubinacci S, Ribeiro DM, Hofmeister RJ, Delaneau O. Rubinacci S, et al. Nat Genet. 2021 Jan;53(1):120-126. doi: 10.1038/s41588-020-00756-0. Epub 2021 Jan 7. Nat Genet. 2021. PMID: 33414550
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous