The role of miR-31 and its target gene SATB2 in cancer-associated fibroblasts - PubMed (original) (raw)
The role of miR-31 and its target gene SATB2 in cancer-associated fibroblasts
Olga Aprelikova et al. Cell Cycle. 2010.
Abstract
It is well established that there is a dynamic relationship between the expanding tumor and the host surrounding tissue. Cancer-associated fibroblasts (CAFs), the most common cellular population found in the tumor microenvironment, supporting tumor growth and dissemination. Here, we set out to determine the factors that may be involved in dramatic alteration of gene expression pattern in CAFs, focusing on microRNA and transcriptional regulators. We established matched pairs of human CAFs isolated from endometrial cancer and normal endometrial fibroblasts. MicroRNA and mRNA analyses identified differential expression of 11 microRNAs, with miR-31 being the most downregulated microRNA in CAFs (p = 0.007). We examined several putative miR-31 target genes identified by microarray analysis and demonstrated that miR-31 directly targets the homeobox gene SATB2, which is responsible for chromatin remodeling and regulation of gene expression, and was significantly elevated in CAFs. The functional relevance of miR-31 and SATB2 were tested in in vitro models of endometrial cancer. Overexpression of miR-31 significantly impaired the ability of CAFs to stimulate tumor cell migration and invasion, without affecting tumor cell proliferation. Genetic manipulation of SATB2 levels in normal fibroblasts or CAFs showed that, reciprocally to miR-31, SATB2 increased tumor cell migration and invasion, while knockdown of endogenous SATB2 in CAFs reversed this phenotype. Introduction of SATB2 into normal fibroblasts stimulated expression of a number of genes involved in cell invasion, migration and scattering. These findings provide new insights into tumor-stroma interaction and document that miR-31 and its target gene SATB2, are involved in regulation of tumor cell motility.
Figures
Figure 1
Characterization of fibroblast cell lines. (A) Clinicopathologic data of the samples used to produce fibroblasts. (B) Western blot analysis of fibroblasts (CAFs and NFs) with anti-Fibroblast Specific Protein 1 (FSP1) and anti-Fibroblast Activation Protein (FAP). N, NF; C, CAF. (C) CAFs stimulate growth of endometrial cancer cell line (EC1) compared to normal fibroblasts in co-culture experiments. (D and E) Conditioned media from CAFs stimulate EC1 cells matrigel invasion (D) and migration (E). Values in (C–E) represent average numbers for five pairs of fibroblasts ± SEM. (F) Images of mice 50 days after injection with EC1-luc cells co-mingled with either NFs or CAFs. (G) Quantification of tumor burden in mice injected with EC1-luc alone or in combination with either mouse embryo fibroblasts (MEFs), endometrial NFs or endometrial CAFs.
Figure 2
MicroRNA analysis of CAFs. (A) List of microRNAs differentially expressed in CAFs relative to normal fibroblasts. *The same cluster of microRNAs. #The same family of microRNAs. (B) The microRNAs recognized by the probes representing non-human species on the microarrays. (C) Stem-loop RT-PCR validations of microRNAs differentially expressed in CAFs. Black bars, NF; white bars, CAFs. (D) MicroRNA's predicted target enrichment.
Figure 3
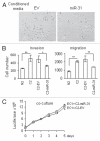
MiR-31 expression in fibroblasts decreases tumor cell migration and invasion. (A) Photographs depict the EC1 cancer cells migration towards the media preconditioned by CAFs stably transduced with miR-31 or empty vector (EV) lentiviral constructs in Transwell migration assays. (B) Quantification of EC1 cell migration or matrigel invasion is presented as mean ± SEM. Experiments were performed in triplicate and repeated at least three times using CAFs from two different patients. Representative experiment is shown. p values were obtained by paired t-test (*p < 0.05, **p < 0.01). (C) miR-31-transduced CAFs do not affect EC1 cell growth in co-culture experiments.
Figure 4
MiR-31-predicted target genes showed differential expression in CAFs. (A) Genes that matched TargetScan predicted genes in the list of genes generated by Significance Analysis of Microarray (SAM) method. Fold induction shows the ratio of expression in CAFs vs. normal fibroblasts. (B) Quantitative RT-PCR validations of SATB2 gene expression in ten pairs of fibroblasts. Black bars, NF; white bars, CAFs. Expression values for CAFs are presented relative to their respective NFs. (C) Western blot analysis of SATB2 protein expression.
Figure 5
SATB2 is a direct target of miR-31. (A) Schematic representation of SATB2 mRNA and the positions of two predicted miR-31 binding sites (S1 and S2) in the 3′UTR. (B) Luciferase activity in HeLa cells after transfection with 1.9 kb SATB2 3′UTR reporter construct containing two binding sites and miR-31 (left part). Ctr, non-targeting control; EV, empty pMIR-Report vector. MiR-31 targets each of the binding sites, S1 (middle part) and S2 (right part) when cloned separately in the pMIR-Report vector. The effect of miR-31 was eliminated by mutations in its binding sites. The experiments were repeated at least three times with similar results, and the representative experiment is shown. (C) Quantitative RT-PCR of SATB2 transcript after overexpression of miR-31 or non-targeting control in CAFs. MiR-148a has been used as additional negative control. The results are mean values of relative mRNA levels normalized to beta-actin ± SEM. From at least three experiments. p-values were obtained by paired t-tests (*p < 0.01). (D) Western blot analysis of SATB2 protein downregulation in five pairs of CAFs after overexpression of miR-31. Endometrial CAFs were transfected with non-targeting control or miR-31 mimic, and three days later cells were collected for western blotting.
Figure 6
SATB2 stimulates endometrial cancer cell migration and invasion. Transwell migration and matrigel invasion experiments were performed using normal fibroblasts (N) transduced with SATB2 lentiviral construct. (A) Microphotographs show endometrial cancer EC1 cells migration towards media pre-conditioned with normal fibroblasts expressing empty vector (EV) control or SATB2. (B) Quantification of EC1 cells migration and invasion experiments were performed as in Figure 4. (C) Co-culture of luciferase-labeled EC1 cells with normal fibroblasts expressing SATB2 or empty vector. (D) Knockdown of SATB2 in CAFs decrease their ability to stimulate EC1 cell migration. NS, non-silencing control. *p = 0.015. (E) Quantitative RT-PCR of SATB2 knockdown by stable expression of lentiviral vector with shSATB2. (F) Western blot analysis of SATB2 protein in CAFs overexpressing shSATB2 or normal fibroblasts with ectopic expression of SATB2 protein. N2 and C2, normal fibroblasts and CAFs, respectively, used for SATB2 knock-down. The last part shows endogenous levels of SATB2 in patient 4, which is comparable with the levels of SATB2 protein overexpressed in normal fibroblasts (patient 2). Same amounts of nuclear lysate were run in the same gel and exposed for the same time.
Figure 7
Expression of SATB2 in normal endometrial fibroblasts induces genes involved in cellular motility. (A) A majority of genes involved in paracrine signaling by fibroblasts (localized in extracellular space or plasma membrane) upregulated by SATB2 and the top network, designated as “cellular movement,” is presented. (B) Top five molecular and cellular functions enriched in SATB2 expressed normal fibroblasts. All genes with more than two-fold up or downregulation by SATB2 were used to assess the SATB2 functions.
Comment in
- MicroRNAs are invading the tumor microenvironment: Fibroblast microRNAs regulate tumor cell motility and invasiveness.
Shurin MR. Shurin MR. Cell Cycle. 2010 Nov 15;9(22):4430-1. doi: 10.4161/cc.9.22.13822. Epub 2010 Nov 15. Cell Cycle. 2010. PMID: 21088483 No abstract available. - miR-31 in cancer: location matters.
Stuelten CH, Salomon DS. Stuelten CH, et al. Cell Cycle. 2010 Dec 1;9(23):4608-9. doi: 10.4161/cc.9.23.13928. Cell Cycle. 2010. PMID: 21260945 No abstract available.
Similar articles
- Elevated microRNA-31 expression regulates colorectal cancer progression by repressing its target gene SATB2.
Yang MH, Yu J, Chen N, Wang XY, Liu XY, Wang S, Ding YQ. Yang MH, et al. PLoS One. 2013 Dec 30;8(12):e85353. doi: 10.1371/journal.pone.0085353. eCollection 2013. PLoS One. 2013. PMID: 24386467 Free PMC article. - miR-101 represses lung cancer by inhibiting interaction of fibroblasts and cancer cells by down-regulating CXCL12.
Zhang J, Liu J, Liu Y, Wu W, Li X, Wu Y, Chen H, Zhang K, Gu L. Zhang J, et al. Biomed Pharmacother. 2015 Aug;74:215-21. doi: 10.1016/j.biopha.2015.08.013. Epub 2015 Aug 28. Biomed Pharmacother. 2015. PMID: 26349988 - MiR-34a inhibits the proliferation, migration, and invasion of oral squamous cell carcinoma by directly targeting SATB2.
Ge X, Gao J, Sun QW, Wang CX, Deng W, Mao GY, Li HQ, Guo SS, Cheng J, Wu YN, Ye JH. Ge X, et al. J Cell Physiol. 2020 May;235(5):4856-4864. doi: 10.1002/jcp.29363. Epub 2019 Oct 29. J Cell Physiol. 2020. PMID: 31663131 - Deregulation of SATB2 in carcinogenesis with emphasis on miRNA-mediated control.
Chen QY, Des Marais T, Costa M. Chen QY, et al. Carcinogenesis. 2019 May 14;40(3):393-402. doi: 10.1093/carcin/bgz020. Carcinogenesis. 2019. PMID: 30916759 Free PMC article. Review. - SATB1 and 2 in colorectal cancer.
Brocato J, Costa M. Brocato J, et al. Carcinogenesis. 2015 Feb;36(2):186-91. doi: 10.1093/carcin/bgu322. Epub 2014 Dec 27. Carcinogenesis. 2015. PMID: 25543122 Free PMC article. Review.
Cited by
- Melanoma miRNA trafficking controls tumour primary niche formation.
Dror S, Sander L, Schwartz H, Sheinboim D, Barzilai A, Dishon Y, Apcher S, Golan T, Greenberger S, Barshack I, Malcov H, Zilberberg A, Levin L, Nessling M, Friedmann Y, Igras V, Barzilay O, Vaknine H, Brenner R, Zinger A, Schroeder A, Gonen P, Khaled M, Erez N, Hoheisel JD, Levy C. Dror S, et al. Nat Cell Biol. 2016 Sep;18(9):1006-17. doi: 10.1038/ncb3399. Epub 2016 Aug 22. Nat Cell Biol. 2016. PMID: 27548915 - Non-Coding RNAs: Foes or Friends for Targeting Tumor Microenvironment.
Szymanowska A, Rodriguez-Aguayo C, Lopez-Berestein G, Amero P. Szymanowska A, et al. Noncoding RNA. 2023 Aug 28;9(5):52. doi: 10.3390/ncrna9050052. Noncoding RNA. 2023. PMID: 37736898 Free PMC article. Review. - Elevated microRNA-31 expression regulates colorectal cancer progression by repressing its target gene SATB2.
Yang MH, Yu J, Chen N, Wang XY, Liu XY, Wang S, Ding YQ. Yang MH, et al. PLoS One. 2013 Dec 30;8(12):e85353. doi: 10.1371/journal.pone.0085353. eCollection 2013. PLoS One. 2013. PMID: 24386467 Free PMC article. - Fibroblast heterogeneity in the cancer wound.
Öhlund D, Elyada E, Tuveson D. Öhlund D, et al. J Exp Med. 2014 Jul 28;211(8):1503-23. doi: 10.1084/jem.20140692. J Exp Med. 2014. PMID: 25071162 Free PMC article. Review. - Relationships of PBMC microRNA expression, plasma viral load, and CD4+ T-cell count in HIV-1-infected elite suppressors and viremic patients.
Witwer KW, Watson AK, Blankson JN, Clements JE. Witwer KW, et al. Retrovirology. 2012 Jan 12;9:5. doi: 10.1186/1742-4690-9-5. Retrovirology. 2012. PMID: 22240256 Free PMC article.
References
- Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. - PubMed
- Hu M, Yao J, Cai L, Bachman KE, van den Brule F, Velculescu V, et al. Distinct epigenetic changes in the stromal cells of breast cancers. Nat Genet. 2005;37:899–905. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials