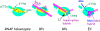Transcriptional control in the prereplicative phase of T4 development - PubMed (original) (raw)
Review
Transcriptional control in the prereplicative phase of T4 development
Deborah M Hinton. Virol J. 2010.
Abstract
Control of transcription is crucial for correct gene expression and orderly development. For many years, bacteriophage T4 has provided a simple model system to investigate mechanisms that regulate this process. Development of T4 requires the transcription of early, middle and late RNAs. Because T4 does not encode its own RNA polymerase, it must redirect the polymerase of its host, E. coli, to the correct class of genes at the correct time. T4 accomplishes this through the action of phage-encoded factors. Here I review recent studies investigating the transcription of T4 prereplicative genes, which are expressed as early and middle transcripts. Early RNAs are generated immediately after infection from T4 promoters that contain excellent recognition sequences for host polymerase. Consequently, the early promoters compete extremely well with host promoters for the available polymerase. T4 early promoter activity is further enhanced by the action of the T4 Alt protein, a component of the phage head that is injected into E. coli along with the phage DNA. Alt modifies Arg265 on one of the two α subunits of RNA polymerase. Although work with host promoters predicts that this modification should decrease promoter activity, transcription from some T4 early promoters increases when RNA polymerase is modified by Alt. Transcription of T4 middle genes begins about 1 minute after infection and proceeds by two pathways: 1) extension of early transcripts into downstream middle genes and 2) activation of T4 middle promoters through a process called sigma appropriation. In this activation, the T4 co-activator AsiA binds to Region 4 of σ⁷⁰, the specificity subunit of RNA polymerase. This binding dramatically remodels this portion of σ⁷⁰, which then allows the T4 activator MotA to also interact with σ⁷⁰. In addition, AsiA restructuring of σ⁷⁰ prevents Region 4 from forming its normal contacts with the -35 region of promoter DNA, which in turn allows MotA to interact with its DNA binding site, a MotA box, centered at the -30 region of middle promoter DNA. T4 sigma appropriation reveals how a specific domain within RNA polymerase can be remolded and then exploited to alter promoter specificity.
Figures
Figure 1
RNAP holoenzyme and the interaction of RNAP with σ 70 -dependent promoters. Structure-based cartoons (left to right) depict RNAP holoenzyme, RPc (closed complex), RPo (open complex), and EC (elongating complex) with σ70 in yellow, core (β,β',α2, and ω) in turquoise, DNA in magenta, and RNA in purple. In holoenzyme, the positions of σ70 Regions 1.1, 2, 3, and 4, the α-CTDs, the β-flap, and the β,β' jaws are identified. In RPc, contact can be made between RNAP and promoter dsDNA elements: two UP elements with each of the α-CTDs, the -35 element with σ70 Region 4, TGn (positions -15 to -13) with σ70 Region 3, and positions -12/-11 of the -10 element with σ70 Region 2. σ70 Region 1.1 lies in the downstream DNA channel formed by portions of β and β' and the β',β' jaws are open. In RPo, unwinding of the DNA and conformational changes within RNAP result in a sharp bend of the DNA into the active site with the formation of the transcription bubble surrounding the start of transcription, the interaction of σ70 Region 2 with nontemplate ssDNA in the -10 element, movement of Region 1.1 from the downstream DNA channel, and contact between the downstream DNA and the β' clamp. In EC, σ70 and the promoter DNA have been released. The newly synthesized RNA remains annealed to the DNA template in the RNA/DNA hybrid as the previously synthesized RNA is extruded through the RNA exit channel past the β-flap.
Figure 2
Comparison of E. coli host, T4 early, and T4 middle promoter sequences. Top, Sequences and positions of host promoter recognition elements for σ70-RNAP (UP, -35, TGn, -10) are shown [20,150]. Below, similar consensus sequences found in T4 early [4] and middle [91] promoters are in black and differences are in red; the MotA box consensus sequence in T4 middle promoters is in green. Spacer lengths between the TGn elements and the -35 elements (host and T4 early) or the MotA box are indicated. W = A or T; R = A or G; Y = C or T, n = any nucleotide; an uppercase letter represents a more highly conserved base.
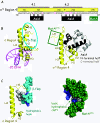
Figure 3
Interaction of σ 70 region 4 with -35 element DNA, the β-flap, AsiA and MotA. A) Sequence of σ70 Region 4 (residues 540-613) with subregions 4.1 and 4.2; the α helices H1 through H5 with a turn (T) between H3 and H4 are shown. Residues of σ70 that interact with the -35 element [25] are colored in magenta. Residues that interact with AsiA [26] or the region that interacts with MotA [97,104] is indicated. B) Structures showing the interaction of T. aquaticus σ Region 4 with -35 element DNA [25] (left, accession # 1KU7) and interaction of σ70 Region 4 with AsiA [26] (right, accession # 1TLH). σ, yellow; DNA, magenta; AsiA, N-terminal half in black, C-terminal half in gray. On the left, the portions of σ that interact with the β-flap (σ residues in and near H1, H2, and H5) are circled in turquoise; on the right, H5, the far C-terminal region of σ70 that interacts with MotA, is in the green square. C) Structures showing the interaction of T. thermophilus σ H5 with the β-flap tip [22] (left, accession # 1IW7) and the structure of MotANTD [94] (right, accession # 1I1S) are shown. On the β-flap (left) and MotANTD (right) structures, hydrophobic residues (L, I, V, or F) and basic residues (K or R) are colored in gray or blue, respectively. The interaction site at the β-flap tip is a hydrophobic hook, while the structure in MotANTD is a hydrophobic cleft.
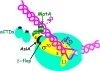
Figure 4
σ appropriation at a T4 middle promoter. Cartoon depicting a model of RPo at a T4 middle promoter (colors as in Fig. 1). Interaction of AsiA with σ70 Region 4 remodels Region 4, preventing its interaction with the β-flap or with the -35 region of the DNA. This interaction then facilitates the interaction of MotANTD with σ70 H5 and MotACTD with the MotA box centered at -30. Protein-DNA interactions at σ70 promoter elements downstream of the MotA box (the TGn and -10 elements) are not significantly affected. ADP-ribosylation of Arg265 on each α-CTD, catalyzed by the T4 Alt and ModA proteins, is denoted by the asterisks. The modification prevents the α subunits from interacting with DNA upstream of the MotA box.
Similar articles
- Transcriptional takeover by sigma appropriation: remodelling of the sigma70 subunit of Escherichia coli RNA polymerase by the bacteriophage T4 activator MotA and co-activator AsiA.
Hinton DM, Pande S, Wais N, Johnson XB, Vuthoori M, Makela A, Hook-Barnard I. Hinton DM, et al. Microbiology (Reading). 2005 Jun;151(Pt 6):1729-1740. doi: 10.1099/mic.0.27972-0. Microbiology (Reading). 2005. PMID: 15941982 Review. - A mutation within the β subunit of Escherichia coli RNA polymerase impairs transcription from bacteriophage T4 middle promoters.
James TD, Cashel M, Hinton DM. James TD, et al. J Bacteriol. 2010 Nov;192(21):5580-7. doi: 10.1128/JB.00338-10. Epub 2010 Aug 20. J Bacteriol. 2010. PMID: 20729353 Free PMC article. - Bacteriophage T4 MotA activator and the β-flap tip of RNA polymerase target the same set of σ70 carboxyl-terminal residues.
Bonocora RP, Decker PK, Glass S, Knipling L, Hinton DM. Bonocora RP, et al. J Biol Chem. 2011 Nov 11;286(45):39290-6. doi: 10.1074/jbc.M111.278762. Epub 2011 Sep 12. J Biol Chem. 2011. PMID: 21911499 Free PMC article. - ADP-ribosylation and early transcription regulation by bacteriophage T4.
Wilkens K, Tiemann B, Bazan F, Rüger W. Wilkens K, et al. Adv Exp Med Biol. 1997;419:71-82. doi: 10.1007/978-1-4419-8632-0_8. Adv Exp Med Biol. 1997. PMID: 9193638 Review.
Cited by
- Inferring bacteriophage infection strategies from genome sequence: analysis of bacteriophage 7-11 and related phages.
Guzina J, Djordjevic M. Guzina J, et al. BMC Evol Biol. 2015;15 Suppl 1(Suppl 1):S1. doi: 10.1186/1471-2148-15-S1-S1. Epub 2015 Feb 2. BMC Evol Biol. 2015. PMID: 25708710 Free PMC article. - Comparative Analysis of 37 Acinetobacter Bacteriophages.
Turner D, Ackermann HW, Kropinski AM, Lavigne R, Sutton JM, Reynolds DM. Turner D, et al. Viruses. 2017 Dec 24;10(1):5. doi: 10.3390/v10010005. Viruses. 2017. PMID: 29295549 Free PMC article. - Structure of Escherichia coli RNA polymerase holoenzyme at last.
Rothman-Denes LB. Rothman-Denes LB. Proc Natl Acad Sci U S A. 2013 Dec 3;110(49):19662-3. doi: 10.1073/pnas.1320604110. Epub 2013 Nov 22. Proc Natl Acad Sci U S A. 2013. PMID: 24272941 Free PMC article. No abstract available. - Context-Dependent Action of Scc4 Reinforces Control of the Type III Secretion System.
Gao L, Cong Y, Plano GV, Rao X, Gisclair LN, Schesser Bartra S, Macnaughtan MA, Shen L. Gao L, et al. J Bacteriol. 2020 Jul 9;202(15):e00132-20. doi: 10.1128/JB.00132-20. Print 2020 Jul 9. J Bacteriol. 2020. PMID: 32424009 Free PMC article. - Diversity, versatility and complexity of bacterial gene regulation mechanisms: opportunities and drawbacks for applications in synthetic biology.
Bervoets I, Charlier D. Bervoets I, et al. FEMS Microbiol Rev. 2019 May 1;43(3):304-339. doi: 10.1093/femsre/fuz001. FEMS Microbiol Rev. 2019. PMID: 30721976 Free PMC article. Review.
References
- Stitt B, Hinton DM. In: Molecular biology of bacteriophage T4. Karam JD, Drake J, Kreuzer KN, Mosig G, Hall D, Eiserling F, Black L, Spicer E, Kutter E, Carlson K, Miller ES, editor. Washington, D.C.: American Society for Microbiology; 1994. Regulation of middle-mode transcription; pp. 142–160.
- Hinton DM, Pande S, Wais N, Johnson XB, Vuthoori M, Makela A, Hook-Barnard I. Transcriptional takeover by sigma appropriation: remodelling of the sigma70 subunit of Escherichia coli RNA polymerase by the bacteriophage T4 activator MotA and co-activator AsiA. Microbiology. 2005;151:1729–1740. doi: 10.1099/mic.0.27972-0. - DOI - PubMed
- Brody E, Rabussay D, Hall D. In: Bacteriophage T4. Mathews CK, Kutter EM, Mosig G, Berget PB, editor. Washington, D. C.: American Society for Microbiology; 1983. Regulation of transcription of prereplicative genes; pp. 174–183.
- Wilkens K, Ruger W. In: Molecular Biology of Bacteriophage T4. Karam JD, Drake JW, Kreuzer KN, Mosig G, Hall DH, Eiserling FA, Black LW, Spicer EK, Kutter E, Carlson K, Miller ES, editor. Washington, D. C.: American Society for Microbiology; 1994. Transcription from early promoters; pp. 132–141.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources