Epigenetic reprogramming in plant and animal development - PubMed (original) (raw)
Review
Epigenetic reprogramming in plant and animal development
Suhua Feng et al. Science. 2010.
Abstract
Epigenetic modifications of the genome are generally stable in somatic cells of multicellular organisms. In germ cells and early embryos, however, epigenetic reprogramming occurs on a genome-wide scale, which includes demethylation of DNA and remodeling of histones and their modifications. The mechanisms of genome-wide erasure of DNA methylation, which involve modifications to 5-methylcytosine and DNA repair, are being unraveled. Epigenetic reprogramming has important roles in imprinting, the natural as well as experimental acquisition of totipotency and pluripotency, control of transposons, and epigenetic inheritance across generations. Small RNAs and the inheritance of histone marks may also contribute to epigenetic inheritance and reprogramming. Reprogramming occurs in flowering plants and in mammals, and the similarities and differences illuminate developmental and reproductive strategies.
Figures
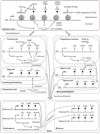
Fig. 1
Model of epigenetic silencing dynamics during Arabidopsis life cycle. In somatic cells, three different mechanisms are responsible for repressing transcription from transposable element (TE), DNA methylation (in all three sequence contexts), histone H3K9 dimethylation (H3K9me2), and histone H3K27 monomethylation (H3K27me1). Methyltransferases and proteins regulating these epigenetic marks are shown in the diagram. See text for details. In the female gametophyte, the central cell is demethylated by DME, which leads to TE activation and upregulation of RdDM. The siRNAs produced from TEs not only direct non-CG methylation in the central cell, but also might travel to egg cell and enhance the silencing of TEs there. In addition, AGO9-associated siRNAs produced in somatic companion cells also contribute to the silencing of TEs in the egg cell. In the male gametophyte, the vegetative nucleus does not express DDM1 and has reduced RdDM, which leads to TE activation and mobilization. A new class of 21-nt siRNAs is produced from TEs in the vegetative nucleus that travels to sperm cells to reinforcing TE silencing. After double fertilization, maternal TEs in the endosperm stay activated and produce PolIV-dependent siRNAs, which could function to silence the paternal TEs in the endosperm. The methylation levels in the embryo are elevated, possibly due to the siRNA signals transmitted from the endosperm. Different shadings indicate the level of DNA methylation, with black being the high, gray being medium, and white being low.
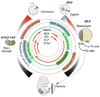
Fig. 2
The two major phases of genome-wide erasure of DNA methylation in the early embryo and in primordial germ cells (PGCs) of the mouse are shown. Thickness of the outer arrows indicates levels of DNA methylation. Red, maternal genome; blue, paternal genome. After fertilization the paternal genome is more rapidly demethylated than the maternal one. During gametogenesis de novo methylation in spermatogenesis occurs earlier than in oogenesis. The inner circle shows factors or candidate factors that are implicated in de novo methylation, the maintenance of methylation, or demethylation, respectively. Solid arrows in the inner circle show at what developmental time these epigenetic regulators are thought to act.
Similar articles
- Epigenetic reprogramming in mammalian development.
Reik W, Dean W, Walter J. Reik W, et al. Science. 2001 Aug 10;293(5532):1089-93. doi: 10.1126/science.1063443. Science. 2001. PMID: 11498579 Review. - Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers.
Seisenberger S, Peat JR, Hore TA, Santos F, Dean W, Reik W. Seisenberger S, et al. Philos Trans R Soc Lond B Biol Sci. 2013 Jan 5;368(1609):20110330. doi: 10.1098/rstb.2011.0330. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23166394 Free PMC article. Review. - Epigenetic reprogramming in mammals.
Morgan HD, Santos F, Green K, Dean W, Reik W. Morgan HD, et al. Hum Mol Genet. 2005 Apr 15;14 Spec No 1:R47-58. doi: 10.1093/hmg/ddi114. Hum Mol Genet. 2005. PMID: 15809273 Review. - DNA Methylation Reprogramming during Mammalian Development.
Zeng Y, Chen T. Zeng Y, et al. Genes (Basel). 2019 Mar 29;10(4):257. doi: 10.3390/genes10040257. Genes (Basel). 2019. PMID: 30934924 Free PMC article. Review. - Transgenerational inheritance: how impacts to the epigenetic and genetic information of parents affect offspring health.
Xavier MJ, Roman SD, Aitken RJ, Nixon B. Xavier MJ, et al. Hum Reprod Update. 2019 Sep 11;25(5):518-540. doi: 10.1093/humupd/dmz017. Hum Reprod Update. 2019. PMID: 31374565
Cited by
- Toward epigenetic and gene regulation models of specific language impairment: looking for links among growth, genes, and impairments.
Rice ML. Rice ML. J Neurodev Disord. 2012 Nov 24;4(1):27. doi: 10.1186/1866-1955-4-27. J Neurodev Disord. 2012. PMID: 23176600 Free PMC article. - AID and Apobec3G haphazard deamination and mutational diversity.
Jaszczur M, Bertram JG, Pham P, Scharff MD, Goodman MF. Jaszczur M, et al. Cell Mol Life Sci. 2013 Sep;70(17):3089-108. doi: 10.1007/s00018-012-1212-1. Epub 2012 Nov 22. Cell Mol Life Sci. 2013. PMID: 23178850 Free PMC article. Review. - Inter-species grafting caused extensive and heritable alterations of DNA methylation in Solanaceae plants.
Wu R, Wang X, Lin Y, Ma Y, Liu G, Yu X, Zhong S, Liu B. Wu R, et al. PLoS One. 2013 Apr 16;8(4):e61995. doi: 10.1371/journal.pone.0061995. Print 2013. PLoS One. 2013. PMID: 23614002 Free PMC article. - Epigenetic regulation in the inner ear and its potential roles in development, protection, and regeneration.
Layman WS, Zuo J. Layman WS, et al. Front Cell Neurosci. 2015 Jan 7;8:446. doi: 10.3389/fncel.2014.00446. eCollection 2014. Front Cell Neurosci. 2015. PMID: 25750614 Free PMC article. Review. - Identification and characterization of paternal-preferentially expressed gene NF-YC8 in maize endosperm.
Mei X, Liu C, Yu T, Liu X, Xu D, Wang J, Wang G, Cai Y. Mei X, et al. Mol Genet Genomics. 2015 Oct;290(5):1819-31. doi: 10.1007/s00438-015-1043-5. Epub 2015 Apr 8. Mol Genet Genomics. 2015. PMID: 25851237
References
- Feng S, et al. Proc Natl Acad Sci U S A. 2010 May 11;107:8689. - PubMed
- Zemach A, McDaniel IE, Silva P, Zilberman D. Science. 2010 May 14;328:916. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 GM060398/GM/NIGMS NIH HHS/United States
- R37 GM060398-10/GM/NIGMS NIH HHS/United States
- BBS/E/B/0000M220/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- HHMI_/Howard Hughes Medical Institute/United States
- G0700098/MRC_/Medical Research Council/United Kingdom
- GM60398/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources