A kinase cascade leading to Rab11-FIP5 controls transcytosis of the polymeric immunoglobulin receptor - PubMed (original) (raw)
. 2010 Dec;12(12):1143-53.
doi: 10.1038/ncb2118. Epub 2010 Oct 31.
Affiliations
- PMID: 21037565
- PMCID: PMC3072784
- DOI: 10.1038/ncb2118
A kinase cascade leading to Rab11-FIP5 controls transcytosis of the polymeric immunoglobulin receptor
Tao Su et al. Nat Cell Biol. 2010 Dec.
Abstract
Polymeric immunoglobulin A (pIgA) transcytosis, mediated by the polymeric immunoglobulin receptor (pIgR), is a central component of mucosal immunity and a model for regulation of polarized epithelial membrane traffic. Binding of pIgA to pIgR stimulates transcytosis in a process requiring Yes, a Src family tyrosine kinase (SFK). We show that Yes directly phosphorylates EGF receptor (EGFR) on liver endosomes. Injection of pIgA into rats induced EGFR phosphorylation. Similarly, in MDCK cells, pIgA treatment significantly increased phosphorylation of EGFR on various sites, subsequently activating extracellular signal-regulated protein kinase (ERK). Furthermore, we find that the Rab11 effector Rab11-FIP5 is a substrate of ERK. Knocking down Yes or Rab11-FIP5, or inhibition of the Yes-EGFR-ERK cascade, decreased pIgA-pIgR transcytosis. Finally, we demonstrate that Rab11-FIP5 phosphorylation by ERK controls Rab11a endosome distribution and pIgA-pIgR transcytosis. Our results reveal a novel Yes-EGFR-ERK-FIP5 signalling network for regulation of pIgA-pIgR transcytosis.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Figures
Figure 1
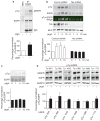
Identification of EGFR as a Yes substrate in rat liver endosomes. (a) A Yes-GTM kinase reaction assay was performed in vitro on endosomal membranes for the indicated times. Each reaction mixture contained rat liver endosomes and [γ-32P]–_N_6(benzyl) ATP, with or without Yes-GTM as indicated. Proteins were separated by SDS–PAGE and 32P-labelled proteins were detected by a phosphorimager. Three major proteins of 170, 46 and 44 K are phosphorylated by Yes-GTM. Boxes indicate separate gels. (b) Quantification of the band intensities in a. (c, d) Kinase reaction assays were performed with or without Yes-GTM, and with PP2 or PP3 (c) or 1-NM-PP1 (d) at the indicated concentrations. Reaction mixtures contained rat liver endosomes and [γ-32P]–_N_6(benzyl) ATP. Proteins were separated by SDS–PAGE and 32P-labelled proteins were detected by a phosphorimager. PP3 is an inactive analogue of PP2. (e) Protein from endosomes was resolved by SDS–PAGE. Total (green) and phosphorylated EGFR (red) were identified by dual-colour infrared immunoblotting using antibodies specific to EGFR and phosphorylated tyrosine (pTyr). The phosphorylated tyrosine band occurs only in the presence of Yes-GTM, precisely co-migrating with EGFR. Uncropped images of blots are shown in Supplementary Information, Fig. S7.
Figure 2
EGFR phosphorylation is induced in rat liver endosomes, and in pIgR-expressing MDCK cells, on pIgA stimulation. (a) EGFR phosphorylation in rats injected with pIgA. Top: endosome fractions, from rats treated with pIgA as indicated, were analysed by immunoprecipitation of EGFR, and immunoblotting with antibodies against phosphorylated tyrosine and EGFR. Antibody heavy chains from immunoprecipitation are indicated at the bottom of the blot. IP; immunoprecipitation, pEGFR; phosphorylated EGFR. Bottom: intensity of the phosphorylated tyrosine bands (normalized to EGFR bands), with or without pIgA treatment. Data are means ± s.e.m. Asterisk indicates P < 0.03, n = 3. (b) Top: MDCK cells expressing pIgR and stably expressing control, scrambled-sequence shRNA or Yes shRNA were treated basolaterally with pIgA for the indicated times. Cells were lysed and proteins were resolved by SDS–PAGE and immunoblotting with antibodies against phosphorylated tyrosine, EGFR and Yes at the indicated times after pIgA treatment (0 min represents control without pIgA treatment). Colour image represents overlay of phosphorylated tyrosine/EGFR signals from infrared immunoblots. Note precise co-migration of phosphorylated tyrosine and the upper EGFR band in dual-colour immunoblots, representing phosphorylated tyrosine–EGFR. MLC; myosin light chain, loading control. Bottom: intensity of the phosphorylated EGFR bands (normalized to EGFR bands) at indicated times after pIgA treatment. Data are means ± s.e.m. Asterisks indicate P < 0.01, compared with control cells at 0 min, n = 4. (c) Top: pIgR-expressing MDCK cells were treated basolaterally with pIgA for the indicated times in the presence of SFK inhibitor (PP2). Cells were lysed at the indicated times after pIgA treatment and proteins were resolved by SDS–PAGE and immunoblotting. Bottom: intensity of the phosphorylated EGFR bands (normalized to EGFR bands), at indicated times after pIgA treatment. Data are means ± s.e.m., n = 4. (d) Top: MDCK cells stably expressing hEGFR and pIgR were treated basolaterally with pIgA for 5 min, as indicated. Lysates were immunoblotted with antibodies specific to EGFR proteins phosphorylated at the tyrosine residues indicated at the top, and antibodies against total EGFR and GAPDH (glyceraldehyde-3-phosphate dehydrogenase, as a control). Bottom: intensity of the phosphorylated EGFR bands (phosphorylated at the indicated residues and normalized to EGFR bands), with or without pIgA treatment. Data are means ± s.e.m. Asterisk indicates P < 0.03, compared with the respective cells not treated with pIgA (n = 4). Uncropped images of blot are shown in Supplementary Information, Fig. S7.
Figure 3
Interaction and co-localization of EGFR, pIgR and Yes. (a) Rat liver endosomes were solubilized and EGFR, pIgR or Yes were immunoprecipitated. The proteins were resolved by SDS–PAGE and co-immunoprecipitation was assessed by immunoblotting. Immunoprecipitations with non-specific rabbit serum (NSS) were performed as a negative control. In the RRC lane, 5 μg of protein was loaded as the input. The intensity of the co-immunoprecipitated protein bands are indicated, compared with the intensity of the immunoprecipitated protein bands (data are means ± s.e.m., n = 4). Boxes represent spliced regions from same gels, to maintain sample order format under all conditions. (b) pIgR, Yes or EGFR were immunoprecipitated from the lysates of MDCK cells expressing hEGFR and pIgR, and co-immunoprecipitation was detected by immunoblotting, as indicated. Immunoblotting of total cell lysates and immunoprecipitation and immunoblotting of non-specific serum (NSS) and was also performed. The intensity of the co-immunoprecipitated protein bands are indicated as a percentage of the intensity of the protein bands in the input lysate (data are means ± s.e.m., n = 3). (c) Monolayers of MDCK cells expressing hEGFR and pIgR were fixed and stained with antibodies against the indicated proteins for immunofluorescence microscopy. Top: Representative images from the sub-apical and lateral (middle) regions of MDCK cell monolayers. Arrows indicate vesicular co-localization of pIgR, Yes and EGFR. White spots in the merged image indicate intracellular co-localization of EGFR, pIgR and Yes. Bottom: representative images from the sub-apical and lateral (middle) regions of MDCK cell monolayers on pIgA treatment. Scale bars, 20 μm. Uncropped images of blots are shown in Supplementary Information, Fig. S7.
Figure 4
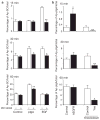
pIgA-stimulated pIgR transcytosis requires EGFR activity. (a) MDCK cells expressing pIgR were labelled with 35S-cysteine and basolaterally treated with pIgA or EGF, with or without EGFR kinase inhibitor (PD153035) for the indicated times. Apically released secretory component (Ap-SC) is represented as a percentage of total labelled pIgR. Data are means ± s.e.m. Asterisk indicates P < 0.05 and double asterisks indicate P < 0.001, compared with cells not treated with PD153035 for the indicated treatment. Control cells with PD153035 treatment, n = 8, all others, n = 6. (b) MDCK cells expressing hEGFR and pIgR, MDCK cells expressing pIgR with Yes knockdown, and their respective controls, were basolaterally treated with biotinylated pIgA for indicated times, and the apically transcytosed pIgA (Ap-pIgA) was measured by ELISA. Data are represented as a percentage of the total pIgA and are means ± s.e.m. Asterisk indicates P < 0.05, double asterisks indicate P < 0.001, compared with respective control cells. Control and hEGFR-expressing cells, n = 8; control and Yes knockdown cells, n = 12.
Figure 5
ERK phosphorylation induced by pIgA treatment is required for pIgA–pIgR transcytosis in MDCK cells expressing pIgR. (a–f) MDCK cells expressing pIgR were treated basolaterally with pIgA for the indicated times. Cells were left untreated (a, control), Yes was knocked down (b), or cells were treated with SFK inhibitor (PP2; c), EGFR inhibitor (PD153035; d) or MEK inhibitor (U0126; e). Top: cell lysates were analysed by immunoblotting with specific phosphorylated ERK (pERK) and ERK antibodies. Bottom: quantification of the phosphorylated ERK band intensities (normalized to the ERK bands). Asterisk (a) indicates P < 0.03, compared with control cells at 0 min, n = 4). (f) Cells were pre-treated for 2 h, and throughout the experiment, with the indicated inhibitors. Control cells were treated with DMSO. Apical pIgA transcytosis of basolaterally applied pIgA was analysed at the indicated times after addition of inhibitor. Data are represented as a percentage of the total pIgA and are means ± s.e.m. Double asterisks indicate P < 0.001, compared with control cells, n = 4).
Figure 6
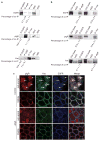
FIP5 phosphorylation is downstream of Yes–EGFR–ERK. (a) Comparison of the amino-acid sequences of FIP5 from the indicated species. A conserved ERK phosphorylation sequence at Ser 188 is indicated. Bottom: schematic representation of the FIP5 protein, indicating region compared at the top. N, amino-terminus; C, carboyxl terminus; RBD, Rab11-binding domain. (b) Immunoblotting of RRC fractions with antibodies against the indicated proteins. Box indicates splicing of bands from same gel. (c) Immunoblotting of the lysates from pIgR-expressing MDCK cells stably expressing FIP5 shRNA and control cells expressing scrambled-sequence shRNA. Duplicate samples are presented. GAPDH; loading control. (d) pIgA transcytosis in pIgR-expressing MDCK cells stably expressing scrambled-sequence shRNA, or FIP5 shRNA. Data are represented as a percentage of the total pIgA and are means ± s.e.m. Asterisks indicate P < 0.001, compared with control cells, n = 4. (e) Expression of FIP5 in MDCK cells. Levels of FIP5 were assessed in parental MDCK cells (left) and in cells overexpressing wild-type human FIP5 or an S188A mutant, both tagged with GFP. GAPDH, loading control. Box indicates splicing of bands from same gel. (f) MDCK cells stably expressing pIgR and GFP–FIP5 (wild type or S188A) were treated with pIgA for the indicated times. Top: cell lysates were immunoprecipitated with antibodies against GFP–FIP5 and then immunoblotted with antibodies specific to GFP–FIP5 and phosphorylated serine (pS). Bottom: intensity of the phosphorylated serine bands (normalized to the intensity of the GFP–FIP5 bands). Data are means ± s.e.m. Single asterisks indicate P < 0.05, double asterisks indicate P < 0.001, n = 4. (g) MDCK cells expressing pIgR and GFP–FIP5 were treated with pIgA for indicated times and with the indicated inhibitors (untreated cells, control). Cell lysates were immunoprecipitated for GFP–FIP5, resolved by SDS–PAGE, and immunoblotted for GFP–FIP5 or phosphorylated serine. Intensity of bands for phosphorylated serine were normalized to the intensity of GFP–FIP5 bands. Data are means ± s.e.m. Asterisks indicate P < 0.05, n = 3. Uncropped images of blots are shown in Supplementary Information, Fig. S7.
Figure 7
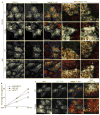
FIP5 Ser 188 phosphorylation regulates Rab11a localization and pIgA–pIgR transcytosis. (a) Filter-grown monolayers of MDCK cells expressing pIgR and stably expressing GFP–FIP5 (wild type or the S188A mutant, green) were immunostained for Rab11a (red) and E-cadherin (E-cad, blue) without (control) or with pIgA stimulation (15 min). Pairs of images on the right are higher-magnification images of boxed areas indicated in images on the left and demonstrate accumulation of Rab11a/FIP5-positive vesicles in the periphery of the sub-apical region of cells expressing GFP–FIP5S188A, as indicated by arrows. (b) pIgA transcytosis assays were performed on parental MDCK cells expressing pIgR or MDCK cells expressing pIgR and GFP–FIP5 (wild type or the S188A mutant). Cells were treated with biotinylated pIgA, which was allowed to accumulate intracellularly, followed by incubation. Transcytosed pIgA was measured at the indicated times. Data are means ± s.e.m. Asterisks indicate P < 0.001, n = 4. (c) MDCK cells expressing pIgR and GFP–FIP5 (green; wild type; top, or the S188A mutant; bottom) were treated basolaterally with biotinylated pIgA, and immunostained for pIgA (red) and F-actin (blue) after 60 min of transcytosis. Yellow, arrowheads indicate overlap of pIgA and wild-type GFP–FIP5 in the centre of the sub-apical region of cells. Images on the right are higher-magnification images of boxed areas indicated in images on the left. Scale bars, 20 μm.
Figure 8
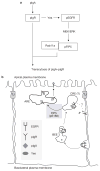
A kinase cascade regulating pIgR transcytosis. (a) A Yes–EGFR–ERK–FIP5 signalling cascade controls pIgA–pIgR transcytosis in epithelial cells. pIgA stimulates pIgR activation, which associates with Yes, and directs phosphorylation of EGFR. Active phosphorylated EGFR, presumably through Ras/Raf, activates MEK/ERK, which in turn phosphorylates FIP5 on Ser 188 (pFIP5). FIP5, phosphorylated on Ser 188, functions with Rab11a to regulate transcytosis of pIgA–pIgR complexes. (b) FIP5 phosphorylation controls polarized distribution of Rab11a and pIgA transcytosis. A schematic representation of how pIgR–Yes–EGFR complexes are internalized and passaged through basolateral early endosomes (BEEs) to sub-apical endosomes, presumably the common recycling endosome (CREs). pIgA causes the disruption of the pIgR–Yes–EGFR complex in endosomes. The EGFR–MEK/ERK cassette phosphorylates FIP5 on Ser 188 (pS188), which controls re-distribution of Rab11a/FIP5 vesicles from the periphery to the centre of the apical region of cells. This may represent the transition from the CRE to the apical recycling endosome (ARE). From here EGFR may be recycled to the basolateral plasma membrane. pIgA–pIgR complexes are delivered to the ARE, from where they are delivered to the apical plasma membrane. Thus the EGFR–MEK/ERK cassette represents an unappreciated regulator of transport through the transcytotic pathway. TJ; tight junctions.
Similar articles
- Rab11-FIP1 and Rab11-FIP5 Regulate pIgR/pIgA Transcytosis through TRIM21-Mediated Polyubiquitination.
Fan X, Zhou D, Zhao B, Sha H, Li M, Li X, Yang J, Yan H. Fan X, et al. Int J Mol Sci. 2021 Sep 28;22(19):10466. doi: 10.3390/ijms221910466. Int J Mol Sci. 2021. PMID: 34638806 Free PMC article. - The SRC family protein tyrosine kinase p62yes controls polymeric IgA transcytosis in vivo.
Luton F, Vergés M, Vaerman JP, Sudol M, Mostov KE. Luton F, et al. Mol Cell. 1999 Oct;4(4):627-32. doi: 10.1016/s1097-2765(00)80213-0. Mol Cell. 1999. PMID: 10549294 - Polymeric IgA binding to the human pIgR elicits intracellular signalling, but fails to stimulate pIgR-transcytosis.
Giffroy D, Courtoy PJ, Vaerman JP. Giffroy D, et al. Scand J Immunol. 2001 Jan;53(1):56-64. doi: 10.1046/j.1365-3083.2001.00843.x. Scand J Immunol. 2001. PMID: 11169207 - Role of Polymeric Immunoglobulin Receptor in IgA and IgM Transcytosis.
Wei H, Wang JY. Wei H, et al. Int J Mol Sci. 2021 Feb 25;22(5):2284. doi: 10.3390/ijms22052284. Int J Mol Sci. 2021. PMID: 33668983 Free PMC article. Review. - The polymeric immunoglobulin receptor: bridging innate and adaptive immune responses at mucosal surfaces.
Kaetzel CS. Kaetzel CS. Immunol Rev. 2005 Aug;206:83-99. doi: 10.1111/j.0105-2896.2005.00278.x. Immunol Rev. 2005. PMID: 16048543 Review.
Cited by
- Extracellular movement of signaling molecules.
Müller P, Schier AF. Müller P, et al. Dev Cell. 2011 Jul 19;21(1):145-58. doi: 10.1016/j.devcel.2011.06.001. Dev Cell. 2011. PMID: 21763615 Free PMC article. Review. - YES oncogenic activity is specified by its SH4 domain and regulates RAS/MAPK signaling in colon carcinoma cells.
Dubois F, Leroy C, Simon V, Benistant C, Roche S. Dubois F, et al. Am J Cancer Res. 2015 May 15;5(6):1972-87. eCollection 2015. Am J Cancer Res. 2015. PMID: 26269757 Free PMC article. - Rab11-FIP1 and Rab11-FIP5 Regulate pIgR/pIgA Transcytosis through TRIM21-Mediated Polyubiquitination.
Fan X, Zhou D, Zhao B, Sha H, Li M, Li X, Yang J, Yan H. Fan X, et al. Int J Mol Sci. 2021 Sep 28;22(19):10466. doi: 10.3390/ijms221910466. Int J Mol Sci. 2021. PMID: 34638806 Free PMC article. - Cooperativity among secretory IgA, the polymeric immunoglobulin receptor, and the gut microbiota promotes host-microbial mutualism.
Kaetzel CS. Kaetzel CS. Immunol Lett. 2014 Dec;162(2 Pt A):10-21. doi: 10.1016/j.imlet.2014.05.008. Epub 2014 May 27. Immunol Lett. 2014. PMID: 24877874 Free PMC article. Review. - Polarized sorting and trafficking in epithelial cells.
Cao X, Surma MA, Simons K. Cao X, et al. Cell Res. 2012 May;22(5):793-805. doi: 10.1038/cr.2012.64. Epub 2012 Apr 24. Cell Res. 2012. PMID: 22525333 Free PMC article. Review.
References
- Mostov KE, Su T, ter Beest M. Polarized epithelial membrane traffic: conservation and plasticity. Nat Cell Biol. 2003;5:287–293. - PubMed
- Rojas R, Apodaca G. Immunoglobulin transport across polarized epithelial cells. Nat Rev Mol Cell Biol. 2002;3:944–956. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK083330/DK/NIDDK NIH HHS/United States
- P30 DK026743/DK/NIDDK NIH HHS/United States
- R01AI25144/AI/NIAID NIH HHS/United States
- R01 AI025144/AI/NIAID NIH HHS/United States
- R01 EB001987-15/EB/NIBIB NIH HHS/United States
- R01 EB001987-16/EB/NIBIB NIH HHS/United States
- NCRR 01614/PHS HHS/United States
- R01DK083330/DK/NIDDK NIH HHS/United States
- R01 EB001987/EB/NIBIB NIH HHS/United States
- R01 DK074398/DK/NIDDK NIH HHS/United States
- R01DK074398/DK/NIDDK NIH HHS/United States
- R01EB001987/EB/NIBIB NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous