Prioritizing genes associated with prostate cancer development - PubMed (original) (raw)
Meta-Analysis
Prioritizing genes associated with prostate cancer development
Ivan P Gorlov et al. BMC Cancer. 2010.
Abstract
Background: The genetic control of prostate cancer development is poorly understood. Large numbers of gene-expression datasets on different aspects of prostate tumorigenesis are available. We used these data to identify and prioritize candidate genes associated with the development of prostate cancer and bone metastases. Our working hypothesis was that combining meta-analyses on different but overlapping steps of prostate tumorigenesis will improve identification of genes associated with prostate cancer development.
Methods: A Z score-based meta-analysis of gene-expression data was used to identify candidate genes associated with prostate cancer development. To put together different datasets, we conducted a meta-analysis on 3 levels that follow the natural history of prostate cancer development. For experimental verification of candidates, we used in silico validation as well as in-house gene-expression data.
Results: Genes with experimental evidence of an association with prostate cancer development were overrepresented among our top candidates. The meta-analysis also identified a considerable number of novel candidate genes with no published evidence of a role in prostate cancer development. Functional annotation identified cytoskeleton, cell adhesion, extracellular matrix, and cell motility as the top functions associated with prostate cancer development. We identified 10 genes--CDC2, CCNA2, IGF1, EGR1, SRF, CTGF, CCL2, CAV1, SMAD4, and AURKA--that form hubs of the interaction network and therefore are likely to be primary drivers of prostate cancer development.
Conclusions: By using this large 3-level meta-analysis of the gene-expression data to identify candidate genes associated with prostate cancer development, we have generated a list of candidate genes that may be a useful resource for researchers studying the molecular mechanisms underlying prostate cancer development.
Figures
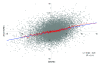
Figure 1
Scatter plot of bone- and non-bone metastasizing cancers. The blue line is a linear regression; the red line is a moving average computed for 100 adjacent genes.
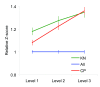
Figure 2
Relative Z score for genes with published evidence of an association with the development of bone metastases based on the KnowledgeNet approach (KN) and for genes from candidate pathways (CP). Relative Z scores were computed as the ratio between the Z score for candidate genes and the overall average Z score. Vertical bars represent standard error (SE).
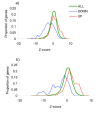
Figure 3
Distributions of Z scores from the third-level analysis. (A) The green line represents the distribution of all Z scores; the blue line, the distribution of Z scores for the genes found to be down-regulated in the study by Chandran et al. [20]; and the red line, the distribution of genes found to be up-regulated in Chandran's study. (B) Distribution of Z scores of genes found to be significantly up- and down-regulated between primary prostate tumor and bone metastases; study by Stanbrough et al., 2006 [21].
Similar articles
- GWAS meets microarray: are the results of genome-wide association studies and gene-expression profiling consistent? Prostate cancer as an example.
Gorlov IP, Gallick GE, Gorlova OY, Amos C, Logothetis CJ. Gorlov IP, et al. PLoS One. 2009 Aug 4;4(8):e6511. doi: 10.1371/journal.pone.0006511. PLoS One. 2009. PMID: 19652704 Free PMC article. - Robust gene network analysis reveals alteration of the STAT5a network as a hallmark of prostate cancer.
Reddy A, Huang CC, Liu H, Delisi C, Nevalainen MT, Szalma S, Bhanot G. Reddy A, et al. Genome Inform. 2010;24:139-53. Genome Inform. 2010. PMID: 22081596 Free PMC article. - Profiling of differential expression of messenger RNA in normal, benign, and metastatic prostate cell lines.
Chakrabarti R, Robles LD, Gibson J, Muroski M. Chakrabarti R, et al. Cancer Genet Cytogenet. 2002 Dec;139(2):115-25. doi: 10.1016/s0165-4608(02)00641-6. Cancer Genet Cytogenet. 2002. PMID: 12550771 - Osteomimetic properties of prostate cancer cells: a hypothesis supporting the predilection of prostate cancer metastasis and growth in the bone environment.
Koeneman KS, Yeung F, Chung LW. Koeneman KS, et al. Prostate. 1999 Jun 1;39(4):246-61. doi: 10.1002/(sici)1097-0045(19990601)39:4<246::aid-pros5>3.0.co;2-u. Prostate. 1999. PMID: 10344214 Review. - Integrative biology of prostate cancer progression.
Tomlins SA, Rubin MA, Chinnaiyan AM. Tomlins SA, et al. Annu Rev Pathol. 2006;1:243-71. doi: 10.1146/annurev.pathol.1.110304.100047. Annu Rev Pathol. 2006. PMID: 18039115 Review.
Cited by
- CTGF (CCN2): a multifaceted mediator in breast cancer progression and therapeutic targeting.
Ghosh P, Dey A, Nandi S, Majumder R, Das S, Mandal M. Ghosh P, et al. Cancer Metastasis Rev. 2025 Feb 13;44(1):32. doi: 10.1007/s10555-025-10248-4. Cancer Metastasis Rev. 2025. PMID: 39945880 Review. - Molecular profiling of prostatic acinar morphogenesis identifies PDCD4 and KLF6 as tissue architecture-specific prognostic markers in prostate cancer.
Li CR, Su JJ, Wang WY, Lee MT, Wang TY, Jiang KY, Li CF, Hsu JM, Chen CK, Chen M, Jiang SS, Weaver VM, Tsai KK. Li CR, et al. Am J Pathol. 2013 Feb;182(2):363-74. doi: 10.1016/j.ajpath.2012.10.024. Epub 2012 Dec 4. Am J Pathol. 2013. PMID: 23219426 Free PMC article. - Androgen-responsive serum response factor target genes regulate prostate cancer cell migration.
Verone AR, Duncan K, Godoy A, Yadav N, Bakin A, Koochekpour S, Jin JP, Heemers HV. Verone AR, et al. Carcinogenesis. 2013 Aug;34(8):1737-46. doi: 10.1093/carcin/bgt126. Epub 2013 Apr 10. Carcinogenesis. 2013. PMID: 23576568 Free PMC article. - miR-888 is an expressed prostatic secretions-derived microRNA that promotes prostate cell growth and migration.
Lewis H, Lance R, Troyer D, Beydoun H, Hadley M, Orians J, Benzine T, Madric K, Semmes OJ, Drake R, Esquela-Kerscher A. Lewis H, et al. Cell Cycle. 2014;13(2):227-39. doi: 10.4161/cc.26984. Epub 2013 Nov 7. Cell Cycle. 2014. PMID: 24200968 Free PMC article. - Identifying Key Genes as Progression Indicators of Prostate Cancer with Castration Resistance Based on Dynamic Network Biomarker Algorithm and Weighted Gene Correlation Network Analysis.
Liu S, Hu Y, Liu F, Jiang Y, Wang H, Wu X, Hu D. Liu S, et al. Biomedicines. 2024 Sep 23;12(9):2157. doi: 10.3390/biomedicines12092157. Biomedicines. 2024. PMID: 39335669 Free PMC article.
References
- Zigeuner R, Droschl N, Tauber V, Rehak P, Langner C. Biologic significance of fascin expression in clear cell renal cell carcinoma: systematic analysis of primary and metastatic tumor tissues using a tissue microarray technique. Urology. 2006;68:518–522. doi: 10.1016/j.urology.2006.03.032. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous