Unfolded Protein Response (UPR) is activated during normal lens development - PubMed (original) (raw)
Unfolded Protein Response (UPR) is activated during normal lens development
Zeynep Firtina et al. Gene Expr Patterns. 2011 Jan-Feb.
Abstract
The lens of the eye is a transparent structure responsible for focusing light onto the retina. It is composed of two morphologically different cell types, epithelial cells found on the anterior surface and the fiber cells that are continuously formed by the differentiation of epithelial cells at the lens equator. The differentiation of an epithelial precursor cell into a fiber cell is associated with a dramatic increase in membrane protein synthesis. How the terminally differentiating fiber cells cope with the increased demand on the endoplasmic reticulum for this membrane protein synthesis is not known. In the present study, we have found evidence of Unfolded Protein Response (UPR) activation during normal lens development and differentiation in the mouse. The ER-resident chaperones, immunoglobulin heavy chain binding protein (BiP) and protein disulfide isomerase (PDI), were expressed at high levels in the newly forming fiber cells of embryonic lenses. These fiber cells also expressed the UPR-associated molecules; XBP1, ATF6, phospho-PERK and ATF4 during embryogenesis. Moreover, spliced XBP1, cleaved ATF6, and phospho-eIF2α were detected in embryonic mouse lenses suggesting that UPR pathways are active in this tissue. These results propose a role for UPR activation in lens fiber cell differentiation during embryogenesis.
Copyright © 2010 Elsevier B.V. All rights reserved.
Figures
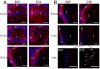
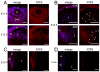
Figure 1. Expression of ER chaperones BiP and PDI in embryonic and adult mouse lenses
Expression of BiP and PDI in WT lenses detected by immunofluorescence. A) At E12.5, both ER chaperones are expressed throughout primary lens fiber cells and lens epithelium (top panels). At E14.5, the expression of BiP and PDI is elevated at the transition zone compared to the central fibers (middle panels) and by E16.5 it becomes mostly confined to the newly forming fiber cells (bottom panels). B) At E18.5, while the expression of both ER chaperones is still high in the newly forming lens fiber cells and lens epithelium, there is only a diffuse staining in central lens fiber cells (top panels). In 3 month old lenses, BiP and PDI expressions are now associated with fiber cells that are getting ready to undergo degradation of nuclei and organelles (middle panels). Bottom panels show higher magnification images of PDI expression in fiber cells. Perinuclear staining in fiber cells that are getting ready to undergo denucleation (white arrow heads) and a dot-like staining at the tips of degrading nuclei (white arrows). Bars, 100 m; e, epithelium; f, fiber cells; tz, transition zone. In all panels blue labels DNA, red labels BiP (left panels) or PDI (right panels).
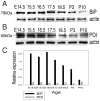
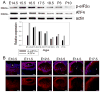
Figure 2. Immunoblot analysis of BiP and PDI expression
A) The major ER chaperone BiP is highly expressed in embryonic lenses. B) Another ER chaperone PDI is also expressed in the lens throughout development; although at lower amounts compared to BiP. Actin was used as a loading control. C) The intensity of the signal on the blots was calculated using Photoshop Histogram Analysis. Relative expression of BiP and PDI normalized to actin is plotted as bar graphs.
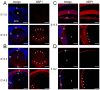
Figure 3. Expression of XBP1 in embryonic and adult lenses
Expression of XBP1 (both spliced and unspliced) in WT lenses detected by immunofluorescence. A) At E11.5, there is no XBP1 expression detected in the lens vesicle (top panels). At E12.5, XBP1 expression becomes apparent throughout lens fiber cells, specifically at the apical and basal tips (bottom panels). B) At E13.5, XBP1 expression is still high throughout lens fiber cell cytoplasm and apical and basal tips (top panels). At E14.5, XBP1 downregulates in lens fiber cells while epithelial XBP1 expression increases (bottom panels). Arrowheads indicate the apical and basal tips of lens fiber cells. C) At E17.5, along with the cornea, lens epithelium shows high XBP1 expression (top panels) while in lens fiber cells, XBP1 expression is observed only in peripheral fiber cells (bottom panels). D) At 3 months, while lens epithelial XBP1 expression stays up (top panels), the expression of XBP1 in fibers is restricted only to the newly forming lens fiber cells (bottom panels). Arrowheads point the newly forming lens fiber cells that express XBP1. Bars, 100 m; c, cornea; e, epithelium; f, fiber cells; lv, lens vesicle; pne, proliferative retinal neuroepithelium; tz, transition zone. In all panels blue labels DNA, red labels XBP1.
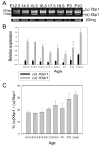
Figure 4. Conventional and real-time RT-PCR analysis of XBP1 splicing
A) Conventional RT-PCR analysis was performed to detect IRE1-induced Xbp1 splicing. There is a basal level of Xbp1 splicing throughout lens development. 2mg was used as a loading control. B) Real-time RT-PCR analysis was performed to quantify Xbp1 expression. The relative expression of total and spliced Xbp1 transcripts normalized to 2mg is plotted as bar graphs. The expression of total Xbp1 at E14.5 is set to 1. C) The bar graph represents the ratio of spliced Xbp1 transcripts to total Xbp1 transcripts during lens development. The spliced Xbp1 transcript represents about %17 of total Xbp1 transcripts at E14.5 and about %32 of total Xbp1 transcripts at 2.5 months.
Figure 5. Immunoblot analysis of XBP1 expression
XBP1(S) protein translated from spliced Xbp1 transcript is found at high levels in embryonic lenses, whereas in postnatal lenses, this protein is not at detectable levels. XBP1(U) protein produced from the unspliced Xbp1 transcript is found at high levels throughout lens development and upregulates postnatally. Actin was used as a loading control. The intensity of the signal on the blots was calculated using Photoshop Histogram Analysis. Relative expression of XBP1(S) and XBP1(U) normalized to actin is plotted as bar graphs.
Figure 6. ATF6 expression in embryonic and adult mouse lenses
Expression of ATF6 (both full length and cleaved) in WT lenses detected by immunofluorescence. A) At E11.5, there are low amounts of ATF6 expression in the posterior lens vesicle cells and the retina (top panels). At E12.5, ATF6 expression is detected at high levels in the fiber cell nuclei (bottom panels). B) At E14.5, there are low amounts of ATF6 expression in the lens epithelium (top panels), while ATF6 is highly expressed by lens fiber cells (bottom panels). Arrowheads indicate the nuclei of lens fiber cells that express ATF6. C) At E18.5, the expression of ATF6 is highest at the transition zone where the newly forming fiber cells are found. D) In 3 month old lenses, ATF6 expression is still detectable in lens epithelium and fiber cells. Bars, 100 m; c, cornea; e, epithelium; f, fiber cells; lv, lens vesicle; pne, proliferative retinal neuroepithelium; tz, transition zone. In all panels blue labels DNA, red labels ATF6.
Figure 7. Immunoblot analysis of ATF6 cleavage
ATF6 protein is cleaved into its nuclear form (ATF6(N)) upon activation. In both embryonic and postnatal normal lenses, ATF6(N) is present. Actin was used as a loading control. The intensity of the signal on the blots was calculated using Photoshop Histogram Analysis. Relative expression of ATF6 and ATF6(N) normalized to actin is plotted as bar graphs.
Figure 8. Expression of p-PERK in embryonic and adult lenses
Expression of p-PERK in WT lenses detected by immunofluorescence. A) At E11.5, there is no p-PERK expression detected in the lens vesicle (top panels). At E12.5, p-PERK expression becomes apparent at the apical tips of lens fiber cells (bottom panels). B) In E14.5 and E16.5 lenses, p-PERK expression is still high at the apical tips of lens fiber cells, specifically at the apical tips of newly forming lens fiber cells. C) At E18.5, p-PERK expression is confined to the peripheral fiber cells. Arrowheads indicate the apical tips of lens fiber cells. D) In 3 months old lenses, p-PERK is expressed by lens fiber cells that are degrading their nuclei. The boxed area indicates the region where p-PERK is expressed. Bars, 100 m; c, cornea; e, epithelium; f, fiber cells; lv, lens vesicle; pne, proliferative retinal neuroepithelium; tz, transition zone. In all panels blue labels DNA, red labels p-PERK.
Figure 9. Expression of ATF4 and p-eIF2 during lens development
A) Western blotting analysis was performed to detect phospho eIF2 (p-eIF2) and ATF4 levels. The levels of p-eIF2 increase around E15.5–E16.5 and downregulate postnatally. Similarly, ATF4 levels are high in embryonic lenses (E14.5–E16.5); however downregulate in postnatal lenses. Actin was used as a loading control. The intensity of the signal on the blots was calculated using Photoshop Histogram Analysis. Relative expression of XBP1(S) and XBP1(U) normalized to actin is plotted as bar graphs. B) Expression of ATF4 in WT lenses detected by immunofluorescence. ATF4 expression is high at E12.5 lens fiber cells but downregulate after E14.5. Bars, 77 m; c, cornea; e, epithelium; f, fiber cells; lv, lens vesicle; tz, transition zone. In all panels blue labels DNA, red labels ATF4.
Similar articles
- Activation of the unfolded protein response in aged human lenses.
Tang HZ, Yang LM. Tang HZ, et al. Mol Med Rep. 2015 Jul;12(1):389-93. doi: 10.3892/mmr.2015.3417. Epub 2015 Mar 4. Mol Med Rep. 2015. PMID: 25739021 - Activation of unfolded protein response in transgenic mouse lenses.
Reneker LW, Chen H, Overbeek PA. Reneker LW, et al. Invest Ophthalmol Vis Sci. 2011 Apr 4;52(5):2100-8. doi: 10.1167/iovs.10-5650. Print 2011 Apr. Invest Ophthalmol Vis Sci. 2011. PMID: 21310900 Free PMC article. - Plasma cell differentiation initiates a limited ER stress response by specifically suppressing the PERK-dependent branch of the unfolded protein response.
Ma Y, Shimizu Y, Mann MJ, Jin Y, Hendershot LM. Ma Y, et al. Cell Stress Chaperones. 2010 May;15(3):281-93. doi: 10.1007/s12192-009-0142-9. Epub 2009 Nov 8. Cell Stress Chaperones. 2010. PMID: 19898960 Free PMC article. - Age-related cataracts: Role of unfolded protein response, Ca2+ mobilization, epigenetic DNA modifications, and loss of Nrf2/Keap1 dependent cytoprotection.
Periyasamy P, Shinohara T. Periyasamy P, et al. Prog Retin Eye Res. 2017 Sep;60:1-19. doi: 10.1016/j.preteyeres.2017.08.003. Epub 2017 Aug 31. Prog Retin Eye Res. 2017. PMID: 28864287 Free PMC article. Review. - Molecular signal networks and regulating mechanisms of the unfolded protein response.
Gong J, Wang XZ, Wang T, Chen JJ, Xie XY, Hu H, Yu F, Liu HL, Jiang XY, Fan HD. Gong J, et al. J Zhejiang Univ Sci B. 2017 Jan.;18(1):1-14. doi: 10.1631/jzus.B1600043. J Zhejiang Univ Sci B. 2017. PMID: 28070992 Free PMC article. Review.
Cited by
- Signaling and Gene Regulatory Networks in Mammalian Lens Development.
Cvekl A, Zhang X. Cvekl A, et al. Trends Genet. 2017 Oct;33(10):677-702. doi: 10.1016/j.tig.2017.08.001. Epub 2017 Aug 31. Trends Genet. 2017. PMID: 28867048 Free PMC article. Review. - A possible connection between reactive oxygen species and the unfolded protein response in lens development: From insight to foresight.
Gao L, Jin N, Ye Z, Ma T, Huang Y, Li H, Du J, Li Z. Gao L, et al. Front Cell Dev Biol. 2022 Sep 21;10:820949. doi: 10.3389/fcell.2022.820949. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36211466 Free PMC article. Review. - The Cataract-linked Mutant Connexin50D47A Causes Endoplasmic Reticulum Stress in Mouse Lenses.
Berthoud VM, Minogue PJ, Lambert PA, Snabb JI, Beyer EC. Berthoud VM, et al. J Biol Chem. 2016 Aug 19;291(34):17569-78. doi: 10.1074/jbc.M115.707950. Epub 2016 Jun 17. J Biol Chem. 2016. PMID: 27317663 Free PMC article. - Modulation of angiogenesis by genetic manipulation of ATF4 in mouse model of oxygen-induced retinopathy [corrected].
Wang X, Wang G, Kunte M, Shinde V, Gorbatyuk M. Wang X, et al. Invest Ophthalmol Vis Sci. 2013 Sep 3;54(9):5995-6002. doi: 10.1167/iovs.13-12117. Invest Ophthalmol Vis Sci. 2013. PMID: 23942974 Free PMC article. - ATF4 May Be Essential for Adaption of the Ocular Lens to Its Avascular Environment.
Xiang J, Pompetti AJ, Faranda AP, Wang Y, Novo SG, Li DW, Duncan MK. Xiang J, et al. Cells. 2023 Nov 16;12(22):2636. doi: 10.3390/cells12222636. Cells. 2023. PMID: 37998373 Free PMC article.
References
- Back SH, Schroder M, Lee K, Zhang K, Kaufman RJ. ER stress signaling by regulated splicing: IRE1/HAC1/XBP1. Methods. 2005;35:395–416. - PubMed
- Bassnett S. The fate of the Golgi apparatus and the endoplasmic reticulum during lens fiber cell differentiation. Invest Ophthalmol Vis Sci. 1995;36:1793–803. - PubMed
- Bassnett S. Three-dimensional reconstruction of cells in the living lens: the relationship between cell length and volume. Exp Eye Res. 2005;81:716–23. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases