The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored - PubMed (original) (raw)
. 2011 Jan;39(Database issue):D561-8.
doi: 10.1093/nar/gkq973. Epub 2010 Nov 2.
Andrea Franceschini, Michael Kuhn, Milan Simonovic, Alexander Roth, Pablo Minguez, Tobias Doerks, Manuel Stark, Jean Muller, Peer Bork, Lars J Jensen, Christian von Mering
Affiliations
- PMID: 21045058
- PMCID: PMC3013807
- DOI: 10.1093/nar/gkq973
The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored
Damian Szklarczyk et al. Nucleic Acids Res. 2011 Jan.
Abstract
An essential prerequisite for any systems-level understanding of cellular functions is to correctly uncover and annotate all functional interactions among proteins in the cell. Toward this goal, remarkable progress has been made in recent years, both in terms of experimental measurements and computational prediction techniques. However, public efforts to collect and present protein interaction information have struggled to keep up with the pace of interaction discovery, partly because protein-protein interaction information can be error-prone and require considerable effort to annotate. Here, we present an update on the online database resource Search Tool for the Retrieval of Interacting Genes (STRING); it provides uniquely comprehensive coverage and ease of access to both experimental as well as predicted interaction information. Interactions in STRING are provided with a confidence score, and accessory information such as protein domains and 3D structures is made available, all within a stable and consistent identifier space. New features in STRING include an interactive network viewer that can cluster networks on demand, updated on-screen previews of structural information including homology models, extensive data updates and strongly improved connectivity and integration with third-party resources. Version 9.0 of STRING covers more than 1100 completely sequenced organisms; the resource can be reached at http://string-db.org.
Figures
Figure 1.
Protein network visualization on the STRING website. The figure shows a composite of two screenshots, illustrating a typical user interaction with STRING (focused on a specific protein network in Saccharomyces cerevisiae). Upon querying the database with four yeast proteins, the resource first reports a raw network consisting of the highest scoring interaction partners (upper left corner). This network can then be rearranged and clustered directly in the browser window revealing tightly connected functional modules (arrow). For each interaction (or protein), additional information is accessible via dedicated pop-up windows; the bottom part of the figure shows an exemplary pop-up with the information regarding a specific yeast protein.
Figure 2.
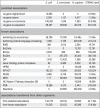
Association counts and data sources. The table shows the number of pair-wise protein–protein associations processed for STRING (version 8.3), listed separately for three important model organisms as well as for the database as a whole. The associations are counted non-directionally, i.e. protein pairs A–B and B–A are counted only once. Identical associations reported by different sources are counted separately under each source, unless they can be traced to the very same publication record and have been imported from primary interaction databases (in case several such databases agree on an interaction, it is arbitrarily counted for only one of them).
Figure 3.
Accessing STRING data from within Cytoscape. Two proteins from Escherichia coli were used as queries for the ‘PSICQUIC Web Service Universal Client’ import-plugin of Cytoscape. Multiple databases have reported hits for these queries (upper left panel); in this case STRING has reported the largest number of hits. The resulting four networks are largely non-overlapping, both in terms of name-spaces as well as in terms of the actual interactors reported. The imported STRING network (right) is shown in detail; it can be used as the basis of further refinement, post-processing and analysis in Cytoscape.
Figure 4.
Projecting third-party data onto the STRING web-surface. STRING provides a consistent name space that encompasses genes, genomes, protein and interaction networks, all of which can be easily searched and browsed. These features can now be employed by external web-resources, via a simple call-back mechanism. External resources can provide cross-links to STRING, together with a call-back address capable of serving a simple text-based interface protocol. At run-time, STRING will then automatically call the external site and project arbitrary ‘payload’ information onto the protein network that is being browsed. The figure shows a fictitious example scenario, served from an in-house test server. As of version 9.0, STRING will also be able to accept protein–protein connections as payload, showing them in a dedicated ‘evidence channel’ distinct from the seven built-in channels. Implementation details are available in the online documentation.
Similar articles
- STRING v9.1: protein-protein interaction networks, with increased coverage and integration.
Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. Franceschini A, et al. Nucleic Acids Res. 2013 Jan;41(Database issue):D808-15. doi: 10.1093/nar/gks1094. Epub 2012 Nov 29. Nucleic Acids Res. 2013. PMID: 23203871 Free PMC article. - STRING 8--a global view on proteins and their functional interactions in 630 organisms.
Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C. Jensen LJ, et al. Nucleic Acids Res. 2009 Jan;37(Database issue):D412-6. doi: 10.1093/nar/gkn760. Epub 2008 Oct 21. Nucleic Acids Res. 2009. PMID: 18940858 Free PMC article. - STRING 7--recent developments in the integration and prediction of protein interactions.
von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Krüger B, Snel B, Bork P. von Mering C, et al. Nucleic Acids Res. 2007 Jan;35(Database issue):D358-62. doi: 10.1093/nar/gkl825. Epub 2006 Nov 10. Nucleic Acids Res. 2007. PMID: 17098935 Free PMC article. - Merging in-silico and in vitro salivary protein complex partners using the STRING database: A tutorial.
Crosara KTB, Moffa EB, Xiao Y, Siqueira WL. Crosara KTB, et al. J Proteomics. 2018 Jan 16;171:87-94. doi: 10.1016/j.jprot.2017.08.002. Epub 2017 Aug 3. J Proteomics. 2018. PMID: 28782718 Review. - GWIDD: a comprehensive resource for genome-wide structural modeling of protein-protein interactions.
Kundrotas PJ, Zhu Z, Vakser IA. Kundrotas PJ, et al. Hum Genomics. 2012 Jul 11;6(1):7. doi: 10.1186/1479-7364-6-7. Hum Genomics. 2012. PMID: 23245398 Free PMC article. Review.
Cited by
- A systemic transcriptome analysis reveals the regulation of neural stem cell maintenance by an E2F1-miRNA feedback loop.
Palm T, Hemmer K, Winter J, Fricke IB, Tarbashevich K, Sadeghi Shakib F, Rudolph IM, Hillje AL, De Luca P, Bahnassawy L, Madel R, Viel T, De Siervi A, Jacobs AH, Diederichs S, Schwamborn JC. Palm T, et al. Nucleic Acids Res. 2013 Apr 1;41(6):3699-712. doi: 10.1093/nar/gkt070. Epub 2013 Feb 8. Nucleic Acids Res. 2013. PMID: 23396440 Free PMC article. - Autworks: a cross-disease network biology application for Autism and related disorders.
Nelson TH, Jung JY, Deluca TF, Hinebaugh BK, St Gabriel KC, Wall DP. Nelson TH, et al. BMC Med Genomics. 2012 Nov 28;5:56. doi: 10.1186/1755-8794-5-56. BMC Med Genomics. 2012. PMID: 23190929 Free PMC article. - Systems biology of pathogen-host interaction: networks of protein-protein interaction within pathogens and pathogen-human interactions in the post-genomic era.
Durmuş Tekir SD, Ülgen KÖ. Durmuş Tekir SD, et al. Biotechnol J. 2013 Jan;8(1):85-96. doi: 10.1002/biot.201200110. Epub 2012 Nov 29. Biotechnol J. 2013. PMID: 23193100 Free PMC article. Review. - Multilevel omic data integration in cancer cell lines: advanced annotation and emergent properties.
Liu Y, Devescovi V, Chen S, Nardini C. Liu Y, et al. BMC Syst Biol. 2013 Feb 19;7:14. doi: 10.1186/1752-0509-7-14. BMC Syst Biol. 2013. PMID: 23418673 Free PMC article. - STRING v9.1: protein-protein interaction networks, with increased coverage and integration.
Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. Franceschini A, et al. Nucleic Acids Res. 2013 Jan;41(Database issue):D808-15. doi: 10.1093/nar/gks1094. Epub 2012 Nov 29. Nucleic Acids Res. 2013. PMID: 23203871 Free PMC article.
References
- Pujol A, Mosca R, Farres J, Aloy P. Unveiling the role of network and systems biology in drug discovery. Trends Pharmacol. Sci. 2010;31:115–123. - PubMed
- Klipp E, Wade RC, Kummer U. Biochemical network-based drug-target prediction. Curr. Opin. Biotechnol. 2010;21:511–516. - PubMed
- Janga SC, Diaz-Mejia JJ, Moreno-Hagelsieb G. Network-based function prediction and interactomics: The case for metabolic enzymes. Metab. Eng. 2010 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous