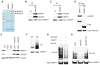DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination - PubMed (original) (raw)
. 2010 Nov 2;3(146):ra80.
doi: 10.1126/scisignal.2001462.
Jing Song, Yong Wang, Yiqing Zhao, Kishore Guda, Shuming Yang, Hung-Ying Kao, Yan Xu, Joseph Willis, Sanford D Markowitz, David Sedwick, Robert M Ewing, Zhenghe Wang
Affiliations
- PMID: 21045206
- PMCID: PMC3116231
- DOI: 10.1126/scisignal.2001462
DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination
Zhanwen Du et al. Sci Signal. 2010.
Abstract
DNA methyltransferase 1 (DNMT1) is the primary enzyme that maintains DNA methylation. We describe a previously unknown mode of regulation of DNMT1 protein stability through the coordinated action of an array of DNMT1-associated proteins. DNMT1 was destabilized by acetylation by the acetyltransferase Tip60, which triggered ubiquitination by the E3 ligase UHRF1, thereby targeting DNMT1 for proteasomal degradation. In contrast, DNMT1 was stabilized by histone deacetylase 1 (HDAC1) and the deubiquitinase HAUSP (herpes virus-associated ubiquitin-specific protease). Analysis of the abundance of DNMT1 and Tip60, as well as the association between HAUSP and DNMT1, suggested that during the cell cycle the initiation of DNMT1 degradation was coordinated with the end of DNA replication and the need for DNMT activity. In human colon cancers, the abundance of DNMT1 correlated with that of HAUSP. HAUSP knockdown rendered colon cancer cells more sensitive to killing by HDAC inhibitors both in tissue culture and in tumor xenograft models. Thus, these studies provide a mechanism-based rationale for the development of HDAC and HAUSP inhibitors for combined use in cancer therapy.
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests. A materials transfer agreement (MTA) is required by Case Western Reserve University for DNMT1, DNMT3B, Tip60 3xFlag knock-in cell lines, the HAUSP knockout cell line, the HDAC1-inducible knockdown cells, and the plasmids generated for this study.
Figures
Fig. 1
HAUSP regulates DNMT1 protein stability. (A) Copurification of DNMT1 with HAUSP. Lysates from DNMT1-3xFlag knock-in (KI) or Mre11-3xFlag KI cells were immunoprecipitated with antibody-conjugated beads against Flag, resolved by SDS–polyacrylamide gel electrophoresis (SDS-PAGE), and stained with GelCode Blue Reagent. (B and C) DNMT1 interacts with HAUSP. RKO cell lysates were immunoprecipitated (IP) with control immunoglobulin G (IgG), antibody against HAUSP or against DNMT1, and then blotted with the indicated antibodies. (D) HAUSP does not interact with DNMT 3b. Cell lysates from DNMT1 and DNMT 3b 3xFlag-tagged KI cells were immunoprecipitated with antibody against Flag and blotted with antibody against HAUSP. (E) DNMT1 abundance is decreased in HAUSP knockout (KO) cells. Cell lysates from DLD1 parental [wild-type (WT)], KO, or KO cells ectopically expressing HAUSP were blotted with the indicated antibodies. KO1 and KO2 are two independently derived HAUSP KO clones. (F) DNMT1 ubiquitination is increased in HAUSP KO cells. HAUSP WT and KO DLD1 cells were treated with 5 μM MG132 for 6 hours. DNMT1 immunoprecipitates were blotted with an antibody against ubiquitin (Ub). (G) HAUSP deubiquitinates DNMT1 in cells. HEK 293 cells were transfected with the indicated plasmids and treated with MG132. DNMT1 immunoprecipitates were blotted with an antibody against HA to detect ubiquitinated DNMT1. (H) HAUSP deubiquitinates DNMT1 in vitro. Ubiquitinated DNMT1 proteins were immunoprecipitated from HEK 293 cells. Equal amounts of recombinant HAUSP or active-site mutant (HAUSP C223S) proteins were added to the immunocomplexes. Western blots were performed to detect either ubiquitinated or total DNMT1 proteins.
Fig. 2
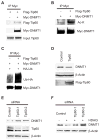
Inhibition of HDAC1-mediated DNMT1 deacetylation promotes DNMT1 proteasomal degradation. (A) Knockout of HAUSP potentiates HDAC inhibitor (HDACi)–induced DNMT1 degradation. Parental or HAUSP KO DLD1 cells were treated or not with 5 μM HDACi MS-275 for 72 hours and cell lysates were blotted with the indicated antibodies. (B) HDAC inhibition induces DNMT1 ubiquitination. HAUSP WT or KO cells were treated with or without HDACi for 24 hours and MG132 for 12 hours before being harvested to make cell lysates. DNMT1 immunoprecipitates were blotted with an antibody against ubiquitin. Because the abundance of DNMT1 in the HAUSP KO cells is lower than in WT cells, more KO cells were used than WT cells to obtain equal amounts of precipitated DNMT1 proteins. (C) DNMT1 is acetylated after HDACi treatment. DNMT1 immunoprecipitates from cells treated with HDACi were blotted with an antibody against acetylated lysine (Ac-K). (D) A DNMT1 acetylation site mutant is resistant to HDACi-induced degradation. HEK 293 cells were transfected with WT DNMT1 or a DNMT1 mutant lacking four known acetylation sites (K173R, K1113R, K115R, and K117R) and treated with MS-275 for 48 hours and with CHX for 24 hours. Cell lysates were blotted with the indicated antibodies. (E) Knockdown of HDAC1 decreases the abundance of DNMT1. RKO cells were treated with the indicated concentration of doxycycline (Dox) for 48 hours to induce expression of an shRNA directed against HDAC1. Western blots were performed with the indicated antibodies. (F) Knockdown of HDAC1 leads to increased acetylation of DNMT1. RKO cells expressing an inducible HDAC1 shRNA were treated with or without Dox (4 μg/ml) for 36 hours and then with MG132 for 12 hours. DNMT1 immunoprecipitates were blotted with an antibody against Ac-K. Cell lysates were also blotted with antibodies against HDAC1 and β-actin. See fig. S11 for bar graphs and statistical analyses.
Fig. 3
Acetylation of DNMT1 by Tip60 promotes its degradation. (A) DNMT1 interacts with Tip60. (B) Tip60 acetylates DNMT1. (C) Overexpression of Tip60 leads to increased ubiquitination of DNMT1. Antibody against Myc immunoprecipitates from HEK 293 cells transfected with the indicated plasmids were immunoblotted with antibody against Flag to detect Tip60 (A), antibody against acetylated lysine to detect DNMT1 acetylation (B), or antibody against HA to detect ubiquitin (C). (D) Over-expression of Tip60 reduced the abundance of endogenous DNMT1. HEK 293 cells were transfected with either control plasmid or Tip60. Cell lysates were blotted with antibody against DNMT1 or antibody against Flag to detect Tip60. β-Actin was used as the loading control. (E) Knockdown of Tip60 leads to increased abundance of endogenous DNMT1. Tip60 Flag-tagged knock-in HCT116 cells were transfected with control siRNA or two independent siRNAs against Tip60. Western blots were used to quantify DNMT1 and Tip60 with antibodies against DNMT1 and Flag, respectively. (F) Knockdown of Tip60 blocks HDACi-induced DNMT1 degradation. Cells were transfected with control siRNA or siRNAs against Tip60 and treated with or without MS-275. Western blots were performed with the indicated antibodies. See fig. S11 for bar graphs and statistical analyses.
Fig. 4
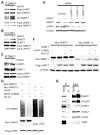
The E3 ligase UHRF1 ubiquitinates DNMT1. (A) HDAC inhibition enhances DNMT1 interaction with UHRF1. HEK 293 cells were transfected with plasmids expressing Myc-DNMT1 and Flag-UHRF1 and treated with or without MS-275 for 24 hours. Myc-DNMT1 immunoprecipitates were blotted with the indicated antibodies. (B and C) HDAC inhibition enhances the interaction of endogenous DNMT1 and UHRF1. Cells were treated with or without MS-275 and UHRF1 (B) or DNMT1 (C) immunoprecipitates were blotted with the indicated antibodies. (D) UHRF1 ubiquitinates DNMT1. HEK 293 cells were transfected with the indicated plasmids. Antibodies against Myc immunoprecipitates were blotted with antibody against HA to detect ubiquitinated DNMT1. Myc-DNMT1Δ, DNMT1 mutant lacking the HAUSP-interacting domain. UHRF1ΔRING, UHRF1 with a RING domain deletion. (E) Knockdown of UHRF1 blocks HDACi-induced DNMT1 degradation. HEK 293 cells were transfected with control siRNA or siRNAs against UHRF1 and treated with or without MS-275. Western blotting was performed with the indicated antibodies. (F) Overexpression of UHRF1 leads to degradation of a DNMT1 mutant lacking the HAUSP-interacting domain (DNMT1Δ). Full-length DNMT1 or DNMT1Δ was cotransfected into HEK 293 cells with the indicated expression vectors. Cell lysates were blotted with the indicated antibodies. (G) DNMT1, HAUSP, UHRF1, HDAC1, and PCNA associate with Tip60. Flag-tagged Tip60 immunoprecipitates were blotted with the indicated antibodies. See fig. S11 for bar graphs and statistical analyses.
Fig. 5
The abundance of DNMT1 correlates with that of HAUSP in colon cancer cells. (A) DNMT1 and HAUSP IHC staining of two representative tumors. Formalin-fixed paraffin-embedded sections of colon adenocarcinomas were stained with antibodies against HAUSP and DNMT1. (B) Correlation of IHC intensity between DNMT1 and HAUSP staining in 10 colon carcinomas (_R_2 = 0.84). Ten low-grade, moderately differentiated adenocarcinomas were examined and the intensity of DNMT1 staining was plotted against the intensity of HAUSP staining for each tumor.
Fig. 6
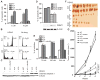
HAUSP KO cells are more sensitive to HDACi-induced apoptosis. (A) HDAC inhibition induces apoptosisinHAUSPKOcells. HAUSP WT or KO cells were treated with or without MS-275 at the indicated concentration for 72 hours, then fixed and stained with propidium iodide. Flow cytometric analyses were used to profile sub-G1, G1, and G2-M cell populations. Apoptotic cells were quantified after the indicated clones were treated with either 5 or 10 μM MS-275. The means and SDs of three independent experiments were plotted (*P < 0.001, t test). (B) HDAC inhibition induces apoptosis in HAUSP KO cells but leads to G2-M arrest in WT cells. Cell cycle profiles of HAUSP WT or KO cells that were treated or not with 5 μM MS-275. (C) HDAC inhibition increases the abundance of apoptotic cell markers. The indicated cells were treated with or without MS-275 for 72 hours. Cell lysates were blotted with antibodies against cleaved caspase 3 and β-actin. (D) Ectopic over-expression of DNMT1 in HAUSP KO cells suppresses apoptosis. HAUSP KO clones or HAUSP KO cells inducibly overexpressing DNMT1 were treated with 10 μMMS-275. Apoptotic cell populations were quantified by fluorescence-activated cell sorting (FACS) analyses (*P < 0.001, t test). Cell lysates from these cells were blotted with the indicated antibodies. (E) HDAC inhibition arrests the growth of HAUSP KO cells. DLD1, HAUSP KO, and KO cells ectopically expressing HAUSP were treated with the indicated concentration of MS-275 for 4 days. Cell numbers were determined and data from eight replicates were plotted (**P < 0.001, t test). (F and G) HDACi inhibits tumor xenograft formation of HAUSPKO cells. Athymic nude mice (five in each group) were injected subcutaneously and bilaterally with cells of the indicated genotypes. Mice were treated with or without MS-275 at 15 mg/kg for 4 weeks. Tumors were harvested and photographed (F). Tumor sizes of the indicated groups were measured weekly and the average volumes at each time point were plotted (G). MANOVA analyses were performed to determine whether there was an overall difference of the tumor sizes, as well as whether there was a difference in development over time of tumor sizes between the two groups (P < 0.0001).
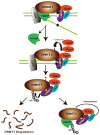
Fig. 7
A model of posttranslational regulation of DNMT1 stability. We propose that DNMT1 physically interacts with HAUSP, Tip60, UHRF1, HDAC1, and PCNA on chromatin. After the completion of DNA methylation in S phase, HAUSP dissociates from DNMT1 to enable DNMT1 degradation. Moreover, increased abundance of Tip60 results in increased acetylation of DNMT1, which in turn triggers the ubiquitination of DNMT1 by UHRF1. This sequence of events results in proteasomal degradation of DNMT1. In contrast, HAUSP and HDAC1 protect DNMT1 from degradation through deubiquitination and deacetylation, respectively.
Similar articles
- Control of DNMT1 abundance in epigenetic inheritance by acetylation, ubiquitylation, and the histone code.
Bronner C. Bronner C. Sci Signal. 2011 Jan 25;4(157):pe3. doi: 10.1126/scisignal.2001764. Sci Signal. 2011. PMID: 21266713 - Usp7 and Uhrf1 control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt1.
Qin W, Leonhardt H, Spada F. Qin W, et al. J Cell Biochem. 2011 Feb;112(2):439-44. doi: 10.1002/jcb.22998. J Cell Biochem. 2011. PMID: 21268065 - Does Wnt/β-catenin pathway contribute to the stability of DNMT1 expression in urological cancer cell lines?
Varol N, Konac E, Bilen CY. Varol N, et al. Exp Biol Med (Maywood). 2015 May;240(5):624-30. doi: 10.1177/1535370214556951. Epub 2014 Oct 27. Exp Biol Med (Maywood). 2015. PMID: 25349215 Free PMC article. - Regulation of maintenance DNA methylation via histone ubiquitylation.
Nishiyama A, Yamaguchi L, Nakanishi M. Nishiyama A, et al. J Biochem. 2016 Jan;159(1):9-15. doi: 10.1093/jb/mvv113. Epub 2015 Nov 20. J Biochem. 2016. PMID: 26590302 Free PMC article. Review. - HAUSP Is a Key Epigenetic Regulator of the Chromatin Effector Proteins.
Abdullah O, Alhosin M. Abdullah O, et al. Genes (Basel). 2021 Dec 24;13(1):42. doi: 10.3390/genes13010042. Genes (Basel). 2021. PMID: 35052383 Free PMC article. Review.
Cited by
- USP7 accelerates FMR1-mediated ferroptosis by facilitating TBK1 ubiquitination and DNMT1 deubiquitination after renal ischemia-reperfusion injury.
Dong B, Ding C, Xiang H, Zheng J, Li X, Xue W, Li Y. Dong B, et al. Inflamm Res. 2022 Dec;71(12):1519-1533. doi: 10.1007/s00011-022-01648-1. Epub 2022 Oct 20. Inflamm Res. 2022. PMID: 36264362 - A Protein Interaction between β-Catenin and Dnmt1 Regulates Wnt Signaling and DNA Methylation in Colorectal Cancer Cells.
Song J, Du Z, Ravasz M, Dong B, Wang Z, Ewing RM. Song J, et al. Mol Cancer Res. 2015 Jun;13(6):969-81. doi: 10.1158/1541-7786.MCR-13-0644. Epub 2015 Mar 9. Mol Cancer Res. 2015. PMID: 25753001 Free PMC article. - Acetylation of ELF5 suppresses breast cancer progression by promoting its degradation and targeting CCND1.
Li X, Li S, Li B, Li Y, Aman S, Xia K, Yang Y, Ahmad B, Wu H. Li X, et al. NPJ Precis Oncol. 2021 Mar 19;5(1):20. doi: 10.1038/s41698-021-00158-3. NPJ Precis Oncol. 2021. PMID: 33742100 Free PMC article. - The stress-responsive gene ATF3 regulates the histone acetyltransferase Tip60.
Cui H, Guo M, Xu D, Ding ZC, Zhou G, Ding HF, Zhang J, Tang Y, Yan C. Cui H, et al. Nat Commun. 2015 Apr 13;6:6752. doi: 10.1038/ncomms7752. Nat Commun. 2015. PMID: 25865756 Free PMC article. - The role of aberrant proteolysis in lymphomagenesis.
Sahasrabuddhe AA, Elenitoba-Johnson KS. Sahasrabuddhe AA, et al. Curr Opin Hematol. 2015 Jul;22(4):369-78. doi: 10.1097/MOH.0000000000000156. Curr Opin Hematol. 2015. PMID: 26049759 Free PMC article. Review.
References
- Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. - PubMed
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. - PubMed
- Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA–(cytosine-5) methyltransferase–PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. - PubMed
- Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DK078965/DK/NIDDK NIH HHS/United States
- R01 DK078965/DK/NIDDK NIH HHS/United States
- R01-CA127590/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- P30 CA043703/CA/NCI NIH HHS/United States
- R01 CA101983/CA/NCI NIH HHS/United States
- R01 HL093269/HL/NHLBI NIH HHS/United States
- R01 HG004722/HG/NHGRI NIH HHS/United States
- R01-CA101983/CA/NCI NIH HHS/United States
- R01-HG004722/HG/NHGRI NIH HHS/United States
- HL093269/HL/NHLBI NIH HHS/United States
- HG004722-02S1/HG/NHGRI NIH HHS/United States
- P30 CA43703/CA/NCI NIH HHS/United States
- R01 CA127590/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous