Alphacoronavirus transmissible gastroenteritis virus nsp1 protein suppresses protein translation in mammalian cells and in cell-free HeLa cell extracts but not in rabbit reticulocyte lysate - PubMed (original) (raw)
Alphacoronavirus transmissible gastroenteritis virus nsp1 protein suppresses protein translation in mammalian cells and in cell-free HeLa cell extracts but not in rabbit reticulocyte lysate
Cheng Huang et al. J Virol. 2011 Jan.
Abstract
The nsp1 protein of transmissible gastroenteritis virus (TGEV), an alphacoronavirus, efficiently suppressed protein synthesis in mammalian cells. Unlike the nsp1 protein of severe acute respiratory syndrome coronavirus, a betacoronavirus, the TGEV nsp1 protein was unable to bind 40S ribosomal subunits or promote host mRNA degradation. TGEV nsp1 also suppressed protein translation in cell-free HeLa cell extract; however, it did not affect translation in rabbit reticulocyte lysate (RRL). Our data suggested that HeLa cell extracts and cultured host cells, but not RRL, contain a host factor(s) that is essential for TGEV nsp1-induced translational suppression.
Figures
FIG. 1.
Effect of TGEV nsp1 on host protein synthesis. (A) HEK293 cells (left) or ST cells (right) were cotransfected with plasmid pRL-SV40 encoding the rLuc reporter gene downstream of the SV40 promoter and one of the following plasmids: pCAGGS-CAT (CAT), pCAGGS-TGEV nsp1-myc (TGEV), pCAGGS-SCoV nsp1-myc (SCoV), and pCAGGS-SCoV nsp1-mt (SCoVmt), expressing the CAT, TGEV nsp1, SCoV nsp1, and SCoV nsp1-mt proteins, respectively. All the expressed proteins, except CAT, had a C-terminal myc epitope tag. At 24 h posttransfection, cell lysates were prepared and subjected to rLuc assay. Error bars show standard deviations (SD) of results from three independent experiments. Cell extracts were also subjected to Western blot analysis by using anti-myc antibody (top) or antiactin antibody (bottom). (B) 293 cells were transfected with _in vitro_-synthesized capped and polyadenylated CAT-myc RNA (lane 1), TGEV nsp1-myc RNA (lane 2), SCoV nsp1-myc RNA (lane 3), and SCoV nsp1-mt-myc RNA (lane 4) using TransIT-mRNA (Mirus). At 1 h post-RNA transfection, cells were mock treated (Act D−) or treated with 4 μg/ml Act D (Act D+) for 7 h. Cells were metabolically labeled with 50 μCi/ml of [35S]methionine for 30 min, and cell lysates were subjected to SDS-PAGE (left). The accumulation of expressed myc-tagged proteins was examined by Western blot analysis using anti-myc antibody (right). (C) ST cells were mock infected (M) or infected (I) with the Purdue strain of TGEV at a multiplicity of infection of 5. Cells were metabolically labeled with 100 μCi/ml of [35S]methionine for 30 min at 4, 6, 8, 10, and 14 h postinfection (hpi). Cell extracts were resolved with 12% SDS-PAGE, and the gels were exposed to X-ray film (autoradiography) or stained with colloidal Coomassie blue (CCB staining). Densitometry analysis was performed to determine the levels of host protein synthesis. The boxes represent the regions of the gel used for densitometry analysis, and the numbers below the lanes of infected cells represent the relative radioactivity compared with that of mock-infected cells at the indicated time postinfection.
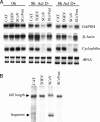
FIG. 2.
Effect of expressed TGEV nsp1 on host mRNA stability. (A) 293 cells were independently transfected with _in vitro_-synthesized capped and polyadenylated CAT-myc RNA (CAT), TGEV nsp1-myc RNA (TGEV), SCoV nsp1-myc RNA (SCoV), and SCoV nsp1-mt-myc RNA (SCoVmt). At 1 h posttransfection, cells were mock treated (Act D−) or treated with 4 μg/ml actinomycin D (Act D+) for 7 h. Total RNAs were extracted at 0 h or 8 h post-RNA transfection and subjected to Northern blot analysis to detect glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA, β-actin mRNA, and cyclophilin mRNA with digoxigenin-labeled antisense riboprobes (11, 15, 22). rRNA, ribosomal RNA (28S [top] and 18S [bottom]). (B) 293 cells were cotransfected with plasmid carrying Ren-EMCV-FF RNA under the control of the SV40 promoter and one of the following plas- mids: pCAGGS-CAT (CAT), pCAGGS-TGEV nsp1-myc (TGEV), pCAGGS-SCoV nsp1-myc (SCoV), and pCAGGS-SCoV nsp1-mt (SCoVmt). At 24 h posttransfection, total RNA was extracted, treated with DNase I, and subjected to Northern blot analysis using a digoxigenin-labeled antisense rLuc riboprobe. Arrows indicate the full-length expressed Ren-EMCV-FF RNA and the RNA fragment generated by SCoV nsp1-induced RNA cleavage.
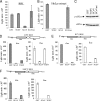
FIG. 3.
Analyses of TGEV nsp1-induced translational suppression in vitro. Capped and polyadenylated GLA mRNA transcripts (0.25 μg) were translated in rabbit reticulocyte lysate (RRL; Promega) for 10 min (A) or HeLa S10 extract for 30 min (B) in the presence of 1 μg of purified GST, TGEV nsp1 (TGEV), SCoV nsp1 (SCoV), or SCoV nsp1-mt (SCoVmt), and rLuc activities were measured. (C) Samples shown in panel B were subjected to Western blot analysis to detect phosphorylated eIF2α (p-eIF2α) and total eIF2α (eIF2α) using anti-phosphorylated eIF2α and anti-eIF2α antibodies (Cell Signaling), respectively. (D) Capped and polyadenylated dicistronic Ren-CrPV-FF RNA (0.25 μg) was translated in HeLa S10 extract for 30 min in the presence of 1 μg of purified GST, SCoV nsp1 (SCoV), or TGEV nsp1 (TGEV). Cap-dependent translation of the rLuc gene and CrPV IRES-driven translation of fLuc were measured by the dual luciferase assay kit (Promega). (E, F) Experiments similar to those shown in panel D were performed, except that Ren-HCV-FF RNA containing HCV IRES (E) and Ren-EMCV-FF RNA containing EMCV IRES (F) were used in place of Ren-CrPV-FF. Error bars show SD of results from three independent experiments.
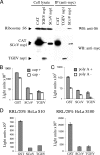
FIG. 4.
Characterization of TGEV nsp1-induced translational suppression. (A) 293 cells were independently transfected with capped and polyadenylated RNA transcripts encoding CAT-myc (CAT), TGEV nsp1-myc (TGEV nsp1), and SCoV nsp1-myc (SCoV nsp1) proteins. At 7 h posttransfection, total cell lysates were prepared and immunoprecipitated with mouse anti-myc antibody (Millipore), followed by stringent washing with high-salt buffer (10 mM HEPES [pH 7.4], 500 mM KCl, 2.5 mM MgCl2 and 1 mM dithiothreitol [DTT]), as described previously (10). Samples were analyzed by Western blotting (WB), using an antibody against S6 protein (anti-S6), which is a component of the 40S ribosome (Cell Signaling). The myc-tagged proteins were detected by using rabbit anti-myc antibody (Cell Signaling) as the primary antibody and horseradish peroxidase (HRP)-conjugated mouse anti-rabbit IgG light-chain-specific antibody as the secondary antibody (Jackson ImmunoResearch) (anti-myc). (B) Capped or uncapped GLA mRNA transcripts were incubated in HeLa S10 extracts for 30 min in the presence of 1 μg of purified GST, SCoV nsp1 (SCoV), or TGEV nsp1 (TGEV). The rLuc activities were measured for capped (gray bars) and uncapped (white bars) GLA mRNA. (C) Capped GLA mRNA transcripts with poly(A) tails (gray bars) or those lacking poly(A) tails (white bars) were incubated in HeLa S10 extracts for 30 min in the presence of purified GST, SCoV nsp1 (SCoV), or TGEV nsp1 (TGEV), and rLuc activities were measured. (D) Capped and polyadenylated GLA mRNA transcripts were incubated in RRL supplemented with 20% HeLa S10 extract (left) or with 20% HeLa S100 extract (right) for 10 min in the presence of 1 μg of purified GST, SCoV nsp1 (SCoV), or TGEV nsp1 (TGEV), and rLuc activities were measured. Error bars show SD of results from three independent experiments.
Similar articles
- A conserved region of nonstructural protein 1 from alphacoronaviruses inhibits host gene expression and is critical for viral virulence.
Shen Z, Wang G, Yang Y, Shi J, Fang L, Li F, Xiao S, Fu ZF, Peng G. Shen Z, et al. J Biol Chem. 2019 Sep 13;294(37):13606-13618. doi: 10.1074/jbc.RA119.009713. Epub 2019 Jul 26. J Biol Chem. 2019. PMID: 31350335 Free PMC article. - Structure of alphacoronavirus transmissible gastroenteritis virus nsp1 has implications for coronavirus nsp1 function and evolution.
Jansson AM. Jansson AM. J Virol. 2013 Mar;87(5):2949-55. doi: 10.1128/JVI.03163-12. Epub 2012 Dec 26. J Virol. 2013. PMID: 23269811 Free PMC article. - A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein.
Kamitani W, Huang C, Narayanan K, Lokugamage KG, Makino S. Kamitani W, et al. Nat Struct Mol Biol. 2009 Nov;16(11):1134-40. doi: 10.1038/nsmb.1680. Epub 2009 Oct 18. Nat Struct Mol Biol. 2009. PMID: 19838190 Free PMC article. - Transmissible gastroenteritis virus infection: a vanishing specter.
Schwegmann-Wessels C, Herrler G. Schwegmann-Wessels C, et al. Dtsch Tierarztl Wochenschr. 2006 Apr;113(4):157-9. Dtsch Tierarztl Wochenschr. 2006. PMID: 16716052 Review. - An overview of immunological and genetic methods for detecting swine coronaviruses, transmissible gastroenteritis virus, and porcine respiratory coronavirus in tissues.
Sirinarumitr T, Paul PS, Halbur PG, Kluge JP. Sirinarumitr T, et al. Adv Exp Med Biol. 1997;412:37-46. doi: 10.1007/978-1-4899-1828-4_4. Adv Exp Med Biol. 1997. PMID: 9191988 Review.
Cited by
- Infectious Bronchitis Coronavirus Limits Interferon Production by Inducing a Host Shutoff That Requires Accessory Protein 5b.
Kint J, Langereis MA, Maier HJ, Britton P, van Kuppeveld FJ, Koumans J, Wiegertjes GF, Forlenza M. Kint J, et al. J Virol. 2016 Jul 27;90(16):7519-7528. doi: 10.1128/JVI.00627-16. Print 2016 Aug 15. J Virol. 2016. PMID: 27279618 Free PMC article. - Nephropathogenic Infectious Bronchitis Virus Mediates Kidney Injury in Chickens via the TLR7/NF-κB Signaling Axis.
Li N, Huang C, Chen W, Li Z, Hu G, Li G, Liu P, Hu R, Zhuang Y, Luo J, Gao X, Guo X. Li N, et al. Front Cell Infect Microbiol. 2022 Mar 23;12:865283. doi: 10.3389/fcimb.2022.865283. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35402297 Free PMC article. - Genomics and pathogenesis of the avian coronavirus infectious bronchitis virus.
Quinteros JA, Noormohammadi AH, Lee SW, Browning GF, Diaz-Méndez A. Quinteros JA, et al. Aust Vet J. 2022 Oct;100(10):496-512. doi: 10.1111/avj.13197. Epub 2022 Aug 17. Aust Vet J. 2022. PMID: 35978541 Free PMC article. Review. - Artesunate induces substantial topological alterations in the SARS-CoV-2 Nsp1 protein structure.
Gurung AB, Ali MA, Lee J, Farah MA, Al-Anazi KM, Al-Hemaid F. Gurung AB, et al. J King Saud Univ Sci. 2022 Feb;34(2):101810. doi: 10.1016/j.jksus.2021.101810. Epub 2022 Jan 3. J King Saud Univ Sci. 2022. PMID: 35002180 Free PMC article. - Translational control of coronaviruses.
de Breyne S, Vindry C, Guillin O, Condé L, Mure F, Gruffat H, Chavatte L, Ohlmann T. de Breyne S, et al. Nucleic Acids Res. 2020 Dec 16;48(22):12502-12522. doi: 10.1093/nar/gkaa1116. Nucleic Acids Res. 2020. PMID: 33264393 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- AI72493/AI/NIAID NIH HHS/United States
- R01 AI026765-20/AI/NIAID NIH HHS/United States
- R01 AI072493/AI/NIAID NIH HHS/United States
- R01 AI026765/AI/NIAID NIH HHS/United States
- AI26765/AI/NIAID NIH HHS/United States
- R56 AI026765/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources