Sequence-specific assembly of FtsK hexamers establishes directional translocation on DNA - PubMed (original) (raw)
Sequence-specific assembly of FtsK hexamers establishes directional translocation on DNA
James E Graham et al. Proc Natl Acad Sci U S A. 2010.
Abstract
FtsK is a homohexameric, RecA-like dsDNA translocase that plays a key role in bacterial chromosome segregation. The FtsK regulatory γ-subdomain determines directionality of translocation through its interaction with specific 8 base pair chromosomal sequences [(KOPS); FtsK Orienting/Polarizing Sequence(s)] that are cooriented with the direction of replication in the chromosome. We use millisecond-resolution ensemble translocation and ATPase assays to analyze the assembly, initiation, and translocation of FtsK. We show that KOPS are used to initiate new translocation events rather than reorient existing ones. By determining kinetic parameters, we show sigmoidal dependences of translocation and ATPase rates on ATP concentration that indicate sequential cooperative coupling of ATP hydrolysis to DNA motion. We also estimate the ATP coupling efficiency of translocation to be 1.63-2.11 bp of dsDNA translocated/ATP hydrolyzed. The data were used to derive a model for the assembly, initiation, and translocation of FtsK hexamers.
Conflict of interest statement
Conflict of interest statement: J.E.G. declares that the preappointed editor, S.C.K., is his present employer, but that there is no scientific or financial conflict of interest.
Figures
Fig. 1.
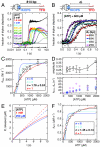
KOPS-dependent and -independent translocation by FtsK measured by triplex displacement. DNA preincubated with FtsK at the indicated monomeric concentrations was rapidly mixed with ATP (final concentration 2 mM). Triplex displacement was monitored by fluorescence and normalized to conditions that yield 100% dissociation. Cartoons show the DNA substrate, with tKOPS as three overlaid blue triangles, TFO as a red line, and FtsK (as side-on hexamer) in gray. Black arrow (A only) shows predicted direction of translocation. (A, B, and C) Concentration-dependent translocation by FtsK on (A) linear tKOPS DNA; (B) linear and negatively supercoiled circular (“circ”) nonspecific DNA (2,732 bp); and (C) linear DNA with nonpermissive tKOPS. (D) Distance dependence of triplex displacement profiles. Profiles are shown for five linear DNAs with different spacings (d T) between tKOPS and TFO. Colored lines show fits to the model (cartoon), assuming one bp represents one  . Kinetic constants are average values (± SD) from fits at each d T. DNA is shown end-on in yellow and ATP binding sites (empty or occupied) in blue. Apparent signal changes before 20 ms resulted from mixing artifacts and were discarded from fitted data.
. Kinetic constants are average values (± SD) from fits at each d T. DNA is shown end-on in yellow and ATP binding sites (empty or occupied) in blue. Apparent signal changes before 20 ms resulted from mixing artifacts and were discarded from fitted data.
Fig. 2.
A six-step model for the assembly of FtsK at KOPS. (A) Two-fork model showing assembly in the absence of ATP, when FtsK is preincubated with DNA and then mixed with ATP (upper branch), and in the presence of ATP, when FtsK is preincubated with ATP and then mixed with DNA and ATP (lower branch). Cartoon as per Fig. 1_D_, with each gray circle representing one FtsK monomer. Kinetic values for assembly are only considered for the lower fork. (B) Effect of preincubating FtsK with DNA in the absence of ATP. Simulations by numerical integration with 1,008 translocation steps, assuming that all DNA molecules are saturated with a hexamer at the start (Left), are compared to the data from Fig. 1_A_ Right. (C) Effect of mixing FtsK and DNA in the presence of saturating ATP. Simulations with 1,008 translocation steps (Left) are compared to equivalent experimental data (Right). FtsK was present at the concentrations indicated and was rapidly mixed with DNA; ATP was present in both syringes at 2 mM. Constants used for both simulations: _k_bind1 = _k_bind2 = _k_bind3 = 1.2 × 109 M-1 s-1; _k_bind4 = _k_bind5 = _k_bind6 = 7.2 × 107 M-1 s-1; _k_off1 = _k_off2 = _k_off3 = 249 s-1; _k_off4 = _k_off5 = _k_off6 = 46.1 s-1; _k_ini = 28 s-1; _k_step = 4,500 s-1; _k_off = 0; and _k_TFO = 46 s-1; _k_bind,ATP → ∞ at 2 mM ATP.
Fig. 3.
ATP coupling in dsDNA translocation by FtsK. (A) Triplex displacement as a function of ATP concentration. A linear DNA with 814 bp spacing between tKOPS and the TFO was preincubated with 50 nM FtsK monomer and rapidly mixed with ATP at the final concentrations shown. (B) Distance-dependence of the triplex displacement profiles at 600 μM ATP. Profiles are shown for four linear DNAs with different spacings (d T) between the tKOPS and TFO. Colored lines show fits by numerical integration as in Fig. 1_D_. (C) Relationship between _k_step and ATP concentration. _k_step values are averages (± SD) derived from multiple DNAs as per (B). Black line is a least squares fit to  where n is the Hill coefficient. Red, cyan, and blue lines are simulations for the n values shown. Residual plots in
where n is the Hill coefficient. Red, cyan, and blue lines are simulations for the n values shown. Residual plots in
Fig. S5
. (D) (Upper) Relationship between displacement amplitude and ATP concentration. No systematic relationship with d T was observed. (Lower) Relationship between _k_ini or _k_TFO and ATP concentration. Points represent values from individual fits for each DNA with no bias to the assignment of _k_ini or _k_TFO (23). (E) Kinetics of phosphate release. DNA was preincubated with FtsK monomer at 50 nM and rapidly mixed with ATP at the concentrations shown. P i release was measured from the fluorescence increase upon binding fluorescently-labeled PBP normalized to a titration of phosphate standard. Dotted lines are linear fits to the first 1 s of the traces to give _k_ATP. (F) Relationship between _k_ATP and ATP concentration. Black line is a least squares fit to Hill equation (C) with red, cyan, and blue lines being fits for the n values shown. Residual plots in
Fig. S5
.
Similar articles
- Single-molecule imaging of DNA curtains reveals mechanisms of KOPS sequence targeting by the DNA translocase FtsK.
Lee JY, Finkelstein IJ, Crozat E, Sherratt DJ, Greene EC. Lee JY, et al. Proc Natl Acad Sci U S A. 2012 Apr 24;109(17):6531-6. doi: 10.1073/pnas.1201613109. Epub 2012 Apr 9. Proc Natl Acad Sci U S A. 2012. PMID: 22493241 Free PMC article. - KOPS-guided DNA translocation by FtsK safeguards Escherichia coli chromosome segregation.
Sivanathan V, Emerson JE, Pages C, Cornet F, Sherratt DJ, Arciszewska LK. Sivanathan V, et al. Mol Microbiol. 2009 Feb;71(4):1031-42. doi: 10.1111/j.1365-2958.2008.06586.x. Epub 2009 Jan 1. Mol Microbiol. 2009. PMID: 19170870 Free PMC article. - Oriented loading of FtsK on KOPS.
Bigot S, Saleh OA, Cornet F, Allemand JF, Barre FX. Bigot S, et al. Nat Struct Mol Biol. 2006 Nov;13(11):1026-8. doi: 10.1038/nsmb1159. Epub 2006 Oct 15. Nat Struct Mol Biol. 2006. PMID: 17041597 - The Escherichia coli DNA translocase FtsK.
Sherratt DJ, Arciszewska LK, Crozat E, Graham JE, Grainge I. Sherratt DJ, et al. Biochem Soc Trans. 2010 Apr;38(2):395-8. doi: 10.1042/BST0380395. Biochem Soc Trans. 2010. PMID: 20298190 Review. - FtsK DNA translocase: the fast motor that knows where it's going.
Crozat E, Grainge I. Crozat E, et al. Chembiochem. 2010 Nov 2;11(16):2232-43. doi: 10.1002/cbic.201000347. Chembiochem. 2010. PMID: 20922738 Review.
Cited by
- Single-molecule imaging of DNA curtains reveals mechanisms of KOPS sequence targeting by the DNA translocase FtsK.
Lee JY, Finkelstein IJ, Crozat E, Sherratt DJ, Greene EC. Lee JY, et al. Proc Natl Acad Sci U S A. 2012 Apr 24;109(17):6531-6. doi: 10.1073/pnas.1201613109. Epub 2012 Apr 9. Proc Natl Acad Sci U S A. 2012. PMID: 22493241 Free PMC article. - Coordinated motion of molecular motors on DNA chains with branch topology.
Lu D, Chen B. Lu D, et al. Acta Mech Sin. 2022;38(3):621225. doi: 10.1007/s10409-021-09045-x. Epub 2022 Feb 16. Acta Mech Sin. 2022. PMID: 35601132 Free PMC article. - Mechanical operation and intersubunit coordination of ring-shaped molecular motors: insights from single-molecule studies.
Liu S, Chistol G, Bustamante C. Liu S, et al. Biophys J. 2014 May 6;106(9):1844-58. doi: 10.1016/j.bpj.2014.03.029. Biophys J. 2014. PMID: 24806916 Free PMC article. - Sequence-dependent catalytic regulation of the SpoIIIE motor activity ensures directionality of DNA translocation.
Chara O, Borges A, Milhiet PE, Nöllmann M, Cattoni DI. Chara O, et al. Sci Rep. 2018 Mar 27;8(1):5254. doi: 10.1038/s41598-018-23400-8. Sci Rep. 2018. PMID: 29588476 Free PMC article. - Active, motor-driven mechanics in a DNA gel.
Bertrand OJ, Fygenson DK, Saleh OA. Bertrand OJ, et al. Proc Natl Acad Sci U S A. 2012 Oct 23;109(43):17342-7. doi: 10.1073/pnas.1208732109. Epub 2012 Oct 8. Proc Natl Acad Sci U S A. 2012. PMID: 23045635 Free PMC article.
References
- Bigot S, et al. FtsK, a literate chromosome segregation machine. Mol Microbiol. 2007;64:1434–1441. - PubMed
- Bigot S, et al. Oriented loading of FtsK on KOPS. Nat Struct Mol Biol. 2006;13:1026–1028. - PubMed
- Ptacin JL, Nöllmann M, Bustamante C, Cozzarelli NR. Identification of the FtsK sequence-recognition domain. Nat Struct Mol Biol. 2006;13:1023–1025. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases