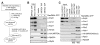A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export - PubMed (original) (raw)
A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export
Thomas Wild et al. PLoS Biol. 2010.
Abstract
The assembly of ribosomal subunits in eukaryotes is a complex, multistep process so far mostly studied in yeast. In S. cerevisiae, more than 200 factors including ribosomal proteins and trans-acting factors are required for the ordered assembly of 40S and 60S ribosomal subunits. To date, only few human homologs of these yeast ribosome synthesis factors have been characterized. Here, we used a systematic RNA interference (RNAi) approach to analyze the contribution of 464 candidate factors to ribosomal subunit biogenesis in human cells. The screen was based on visual readouts, using inducible, fluorescent ribosomal proteins as reporters. By performing computer-based image analysis utilizing supervised machine-learning techniques, we obtained evidence for a functional link of 153 human proteins to ribosome synthesis. Our data show that core features of ribosome assembly are conserved from yeast to human, but differences exist for instance with respect to 60S subunit export. Unexpectedly, our RNAi screen uncovered a requirement for the export receptor Exportin 5 (Exp5) in nuclear export of 60S subunits in human cells. We show that Exp5, like the known 60S exportin Crm1, binds to pre-60S particles in a RanGTP-dependent manner. Interference with either Exp5 or Crm1 function blocks 60S export in both human cells and frog oocytes, whereas 40S export is compromised only upon inhibition of Crm1. Thus, 60S subunit export is dependent on at least two RanGTP-binding exportins in vertebrate cells.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Figure 1. Readouts, phenotypes, and targets used for the systematic analysis of human ribosome biogenesis by RNAi.
(A) Fluorescent images representing the different phenotypic categories used for detecting defects in 40S biogenesis (Rps2-YFP, Enp1 IF) and 60S biogenesis (Rpl29-GFP) after 3 d of RNAi in HeLa cells with the indicated siRNAs (10 nM). (B) Schematic representation of the siRNA target distribution to different protein categories. The list of all targets is given in Table S1.
Figure 2. Ranked high confidence hit list.
Compilation of high confidence hits for the three different readouts, Rps2-YFP, Enp1 IF, and Rpl29-GFP. Rank calculation was as follows: [(hit rate siRNA-X1)−(hit rate negative control)]/[(hit rate positive control)−(hit rate negative control)], where (hit rate negative control) and (hit rate positive control) equals the average hit rate of the two negative or positive controls with the highest hit rate from the same 96-well plate as siRNA-X1. For each hit, the average of the two highest ranked siRNAs is shown. For the Rps2-YFP readout, overall nuclear accumulation (nucleoplasmic and nucleolar) of the reporter was used for hit definition. Note that for “cytoplasmic” Enp1, absolute hit rates are given (ranking could not be performed due to a lack of an appropriate positive control for cytoplasmic accumulation). We set a cutoff to >0.2 for Rps2-YFP, >0.3 for Enp1 IF, and >0.25 for Rpl29-GFP (see Text S1).
Figure 3. Hit distribution and example images of nucleolar and nucleoplasmic accumulation of Rps2-YFP.
(A) Venn diagram of the 153 high confidence hits (Figure 2) and hit enrichment in the different target categories for all readouts (inset). (B) Distribution of high confidence hits to the different categories for each readout. (C) Phenotypic classification of targets with respect to different nuclear 40S biogenesis defects based on nucleolar or nucleoplasmic accumulation of Rps2-YFP. Classification was performed using the ratio of cells with a “nucleoplasmic” phenotype to all cells with a nuclear phenotype (nucleolar and nucleoplasmic). A value of 0 equals 100% of the hit-classified cells have a “nucleolar” phenotype, and a value of 1 equals 100% “nucleoplasmic” hits. Representative images after depletion of selected human SSU processome components (Utp11, Utp3, Imp4, Utp18, Nol14) and NPC components (Nup107, Nup98, Nup54, Nup214, Nup155) are shown.
Figure 4. Requirement of small ribosomal proteins at different stages of 40S biogenesis.
(A) Representative images from Rps2-YFP and Enp1 IF screening analysis after depletion of RPS by RNAi. Positive control: Crm1; negative controls: Allstars (Qiagen) and Rps4Y1 (Y-chromosome encoded). Rps3 depletion is shown as an example of differences detected with the Rps2-YFP (nuclear accumulation) and Enp1 IF (cytoplasmic accumulation) readouts. (B) Rio2 localization after depletion of RPS by RNAi (10 nM siRNAs, 72 h) in the absence or presence of 10 nM leptomycin B (LMB) for 2 h prior to fixation. Endogenous Rio2 was detected by immunofluorescence. Representative images are given for control, Crm1, Rps3, and Rps3A depletion. (C) List of RPS proteins that caused defects in nuclear accumulation of Rio2 upon LMB treatment when downregulated by RNAi in (B). An RPS was defined as a hit if more than 20% of cells were classified as “cytoplasmic” Rio2 after LMB treatment. The average of the two highest scoring siRNAs is given. *For RspA, only 1 siRNA gave a phenotype. (D) RPS were sorted according to the phenotypes detected in the Rps2-YFP, Enp1 IF, and Rio2 IF readouts and grouped with respect to their nuclear or cytoplasmic requirement for human 40S biogenesis. “Nuclear” classified RPS were either detected with Rps2-YFP only (red) or with both the Rps2-YFP and Enp1 IF readouts (orange). Classification of “cytoplasmic” RPS is based on Enp1 IF (yellow), Rio2 IF (+LMB) (brown), or both (green; depletion of these RPS also caused nuclear Rps2-YFP accumulation). FAU was scored as a nuclear Rps2-YFP hit and a cytoplasmic Rio2 IF (+LMB) hit (blue). RPS marked by an asterisk scored positive for one siRNA by at least 7 times the hit rate of negative controls. The yeast RPS classification into four groups (I–IV) is based on and described in the text. For E. coli RPS, primary (1), secondary (2), and tertiary (3) binders of the Nomura assembly map , are listed. The names of yeast and bacterial homologs are given in brackets. FAU and GNBL2L1 are the human ribosomal proteins Rps30 and RACK1, respectively.
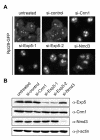
Figure 5. Depletion of Exportin 5 leads to a 60S export defect in human cells.
(A) The Rpl29-GFP cell line was treated with the indicated siRNAs (15 nM) for 72 h. After 52 h of RNAi, Rpl29-GFP expression was induced for 14 h by addition of tetracycline, followed by incubation in tetracycline-free medium for 6 h and subsequent fixation. Images were taken by epifluorescence microscopy. (B) Western blot analysis of extracts from cells derived from the experiment shown in (A). Note that even a slight downregulation of Crm1 results in a prominent and highly reproducible 60S export defect, as also previously described for 40S export .
Figure 6. Exp5 binds specifically to pre-60S particles in a RanGTP-dependent manner.
(A) Schematic representation of the experimental setup. Expression of a tagged bait protein in HEK293 cells was induced by tetracycline. Cells were lysed and the bait protein was affinity-purified by help of its tandem strep/HA-tag using a two-step protocol. Next, the bait protein and associated proteins, bound to anti-HA IgG beads, were incubated with HeLa extracts with or without addition of RanQ69L-GTP. Subsequently, bound proteins were analyzed by Western blotting. (B) Exp5 binds to pre-60S particles in the presence of RanGTP. Pre-60S particles were purified from HEK293 cells bearing an inducible copy of TAP-tagged MRTO4 after 24 h of tetracycline treatment (cell lysate: input MRTO4 TAP). The purified particles were incubated with HeLa cell extract (input HeLa extract) in the absence or presence of 10 µM RanQ69L-GTP. Anti-HA IgG beads were used as a control for non-specific binding of exportins in presence of RanQ69L-GTP. (C) Binding of Exp5 is specific for pre-60S particles. Experiment as in (B) including affinity purification of pre-40S subunits by Dim2 (PNO1) TAP from HEK293 cells bearing an inducible copy of TAP-tagged Dim2. Note that ribosomal particle components are not detected in the dilute input of HEK293 lysates but are highly enriched during purification. Load of input and bound fractions as in (B).
Figure 7. Exp5 is required for nuclear export of 60S subunits in frog oocytes.
X. laevis oocytes were pre-injected in the nucleus with antibodies against Exp5 or Crm1 together with 32P-labeled U3 snRNA and pre-miR-31. To label newly synthesized rRNA, ∼0.5 µCi α-32P-GTP was injected into the cytoplasm 3 h later. After incubation for the indicated times, pools of oocytes were dissected into nuclear (N) and cytoplasmic (C) fractions, and total RNAs extracted. For analyses of large (top panel) or small RNAs (bottom panels), total RNAs were separated by electrophoresis in denaturing 1.2% agarose gels or 8% 7M Urea polyacrylamide gels, respectively. Dried gels were exposed to x-ray film for 20 h (top panel) or 3–4 d (bottom panels). Note the increased accumulation of 28S and 6S(pre-5.8S)(*) rRNAs in the nucleus upon inhibition of pre-60S subunit export. U3 snRNA and pre-miR-31 serve as markers for nuclear injection/proper dissection and Exp5 activity, respectively.
Similar articles
- Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway.
Thomas F, Kutay U. Thomas F, et al. J Cell Sci. 2003 Jun 15;116(Pt 12):2409-19. doi: 10.1242/jcs.00464. Epub 2003 Apr 30. J Cell Sci. 2003. PMID: 12724356 - Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates.
Trotta CR, Lund E, Kahan L, Johnson AW, Dahlberg JE. Trotta CR, et al. EMBO J. 2003 Jun 2;22(11):2841-51. doi: 10.1093/emboj/cdg249. EMBO J. 2003. PMID: 12773398 Free PMC article. - Reengineering ribosome export.
Lo KY, Johnson AW. Lo KY, et al. Mol Biol Cell. 2009 Mar;20(5):1545-54. doi: 10.1091/mbc.e08-10-1000. Epub 2009 Jan 14. Mol Biol Cell. 2009. PMID: 19144820 Free PMC article. - Nuclear export and cytoplasmic maturation of ribosomal subunits.
Zemp I, Kutay U. Zemp I, et al. FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review. - Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast.
Konikkat S, Woolford JL Jr. Konikkat S, et al. Biochem J. 2017 Jan 15;474(2):195-214. doi: 10.1042/BCJ20160516. Biochem J. 2017. PMID: 28062837 Free PMC article. Review.
Cited by
- Probing the mechanisms underlying human diseases in making ribosomes.
Farley KI, Baserga SJ. Farley KI, et al. Biochem Soc Trans. 2016 Aug 15;44(4):1035-44. doi: 10.1042/BST20160064. Biochem Soc Trans. 2016. PMID: 27528749 Free PMC article. Review. - SMALL ORGAN4 Is a Ribosome Biogenesis Factor Involved in 5.8S Ribosomal RNA Maturation.
Micol-Ponce R, Sarmiento-Mañús R, Fontcuberta-Cervera S, Cabezas-Fuster A, de Bures A, Sáez-Vásquez J, Ponce MR. Micol-Ponce R, et al. Plant Physiol. 2020 Dec;184(4):2022-2039. doi: 10.1104/pp.19.01540. Epub 2020 Sep 10. Plant Physiol. 2020. PMID: 32913045 Free PMC article. - Misregulation of Nucleoporins 98 and 96 leads to defects in protein synthesis that promote hallmarks of tumorigenesis.
Pulianmackal AJ, Kanakousaki K, Flegel K, Grushko OG, Gourley E, Rozich E, Buttitta LA. Pulianmackal AJ, et al. Dis Model Mech. 2022 Mar 1;15(3):dmm049234. doi: 10.1242/dmm.049234. Epub 2022 Mar 16. Dis Model Mech. 2022. PMID: 35107131 Free PMC article. - The evolution of the ribosome biogenesis pathway from a yeast perspective.
Ebersberger I, Simm S, Leisegang MS, Schmitzberger P, Mirus O, von Haeseler A, Bohnsack MT, Schleiff E. Ebersberger I, et al. Nucleic Acids Res. 2014 Feb;42(3):1509-23. doi: 10.1093/nar/gkt1137. Epub 2013 Nov 14. Nucleic Acids Res. 2014. PMID: 24234440 Free PMC article. - Ribosome synthesis-unrelated functions of the preribosomal factor Rrp12 in cell cycle progression and the DNA damage response.
Dosil M. Dosil M. Mol Cell Biol. 2011 Jun;31(12):2422-38. doi: 10.1128/MCB.05343-11. Epub 2011 Apr 11. Mol Cell Biol. 2011. PMID: 21482668 Free PMC article.
References
- Mizushima S, Nomura M. Assembly mapping of 30S ribosomal proteins from E. coli. Nature. 1970;226:1214. - PubMed
- Hage A. E, Tollervey D. A surfeit of factors: why is ribosome assembly so much more complicated in eukaryotes than bacteria? RNA Biol. 2004;1:10–15. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous