Use of H19 Gene Regulatory Sequences in DNA-Based Therapy for Pancreatic Cancer - PubMed (original) (raw)
doi: 10.1155/2010/178174. Epub 2010 Oct 28.
V Sorin, Y Fellig, T Birman, A Mizrahi, J Galula, R Abu-Lail, T Shneider, P Ohana, L Buscail, A Hochberg, A Czerniak
Affiliations
- PMID: 21052499
- PMCID: PMC2967839
- DOI: 10.1155/2010/178174
Use of H19 Gene Regulatory Sequences in DNA-Based Therapy for Pancreatic Cancer
V Scaiewicz et al. J Oncol. 2010.
Abstract
Pancreatic cancer is the eighth most common cause of death from cancer in the world, for which palliative treatments are not effective and frequently accompanied by severe side effects. We propose a DNA-based therapy for pancreatic cancer using a nonviral vector, expressing the diphtheria toxin A chain under the control of the H19 gene regulatory sequences. The H19 gene is an oncofetal RNA expressed during embryo development and in several types of cancer. We tested the expression of H19 gene in patients, and found that 65% of human pancreatic tumors analyzed showed moderated to strong expression of the gene. In vitro experiments showed that the vector was effective in reducing Luciferase protein activity on pancreatic carcinoma cell lines. In vivo experiment results revealed tumor growth arrest in different animal models for pancreatic cancer. Differences in tumor size between control and treated groups reached a 75% in the heterotopic model (P = .037) and 50% in the orthotopic model (P = .007). In addition, no visible metastases were found in the treated group of the orthotopic model. These results indicate that the treatment with the vector DTA-H19 might be a viable new therapeutic option for patients with unresectable pancreatic cancer.
Figures
Figure 1
ISH of well-differentiated pancreatic carcinoma from two different patients. (a) Detection of H19 transcript using Dig-labeled H19 LNA-DNA probe by ISH. The arrow points to the positive hybridization signals within the cytoplasm of the cancer cells (10X magnification). (b) Detection of H19 transcript using Dig-labeled H19 LNA-DNA probe by ISH. The arrow points to the positive hybridization signals within the cytoplasm of the cancer cells (20X magnification). Tissue was later stained using Hematoxylin and Eosine.
Figure 2
H19 RNA level in human and hamster pancreatic carcinoma cell lines. (a) Lane 1: 100 bp marker, Lane 2: CRL-1469 cells, Lane 3: PC.1-0 hamster cells. (b) Lane 1: 100 bp marker, Lanes 2–5: CRL-2547, CRL-1687, CRL-2119, and CRL-1997 human pancreatic carcinoma cells, respectively. (c) Lane 1: 100 bp marker; Lane 2: mouse control; Lane 3: PC.1-0 cells cultured under normoxia conditions; Lane 4: PC.1-0 cells cultured under hypoxia condition for 4 hours; Lane 5: tumors generated by PC.1-0 cells injection into nude mice back; Lane 6: negative control, no cDNA is present in the reaction mixture. The upper and the middle panels show the 228 bp and the 454 bp PCR products, respectively, obtained using two different primers (H19-mouse 228 and H19-mouse 454 as described in Supplementary data, Table A). The lower panel indicates the 400 bp _β_-actin internal control.
Figure 3
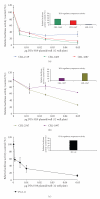
Reduction of Luciferase activity in human and hamster pancreatic carcinoma cell lines due to cotransfection with the DTA-H19 vector. The therapeutic potential of the DTA-H19 vector in different cell lines was measured as a reduction of Luciferase activity. (a) 90000 cells/well were cotransfected with 3 _μ_g of Luc-SV40 plasmid and the indicated amounts of DTA-H19. The luciferase assay was performed 48 hours after transfection. (c) 70000 cells/well were cotransfected with 3 _μ_g of Luc-SV40 plasmid and the indicated concentrations of DTA-H19. Transfection experiments were stopped after 24 hours, and reporter gene activity was assessed. The reduction in the Luc-SV40 activity in the cotransfected cells was compared to cells transfected with Luc-SV40 alone for each cell line. Bar graphs represent the relative Luciferase activity of the H19 promoter when cells were transfected with Luc-H19 plasmid.
Figure 4
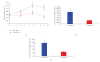
Heterotopic model for pancreatic cancer. (a) Average tumor volume of Luc-H19 group (n = 8) and DTA-H19 group (n = 7) during the experiment using CRL-1469 cells. Treatments were administrated on days 0, 3, and 6 by direct intratumoral injection after subcutaneous implantation of human pancreatic carcinoma cells in the back of nude mice. Three days after the last treatment, the animals were sacrificed. (b) Average of ex vivo tumor volume at the end of the experiment, tumors generated using CRL-1469 cells. (c) Tumor Growth Progression of Luc-H19 (n = 5) and DTA-H19 (n = 5) groups, in experiment using CRL-2547 cell line.
Figure 5
Histopathology analyses and H19 transcript detection in orthotopicaly induced tumors in the pancreas. (a) and (b) Both pictures show poorly differentiated carcinoma. The square in picture (a) delimits the malignant tissue; the circled area in picture (b) corresponds to necrotic interface (X20 original magnification for picture (a), X40 original magnification for picture (b). (c) Interface pancreatic acini (right), neoplasm (left), and desmoplastic reaction (middle) (X10 original magnification). Pictures were taken from paraffin sections and correspond to Giemsa stain. (d)–(f) Total RNA was extracted from frozen pancreatic tumors generated by intracapsular injection of PC.1-0 cells into hamsters pancreas. Lane 1: 100 bp marker, Lane 2: mouse control RNA, Lane 3: RNA extracted from tumor generated in the head of the pancreas, Lanes 4 and 5: RNA extracted from two different tumors generated in the pancreas tail in two different animals. (d) PCR products using primers H19_mouse 454. (e) PCR products using H19-mouse 228 primers. (f) Internal control _β_-actin mRNA.
Figure 6
Effect of DTA-H19 treatment in Tumor Growth Progression and ex vivo tumor size on the orthotopic pancreatic carcinoma in the hamster. (a) Average of tumor growth progression of the DTA-H19 (n = 9) and Luc-H19 (n = 7) treated groups calculated as the ratio of tumor size at the end of the experiment (day 11 after cells implantation) compared to the size before treatment (day 7 after cells implantation). (b) Average of ex vivo tumor volume of the treated (DTA-H19) and control group (Luc-H19) measured after sacrifice (day 11). (c) The left picture corresponds to a tumor treated with DTA-H19 plasmid, and the right picture corresponds to a tumor from the control group.
Figure 7
DTA expression in tumors samples. Total RNA was extracted from frozen pancreatic tumors treated with DTA-H19 plasmid. DNase-treated RNA and nontreated RNA samples were analyzed by RT-PCR for DT-A transcript level. Lane 1: 100 bp marker, Lanes 2 and 4: PCR products from DNase-treated RNA, Lanes 3 and 5: PCR products from nontreated RNA, Lane 6: negative control, no cDNA is present in the reaction mixture. The upper panel shows the 468 bp DT-A product, and the lower panel shows the _β_-actin internal control.
Similar articles
- H19-promoter-targeted therapy combined with gemcitabine in the treatment of pancreatic cancer.
Sorin V, Ohana P, Gallula J, Birman T, Matouk I, Hubert A, Gilon M, Hochberg A, Czerniak A. Sorin V, et al. ISRN Oncol. 2012;2012:351750. doi: 10.5402/2012/351750. Epub 2012 Jun 3. ISRN Oncol. 2012. PMID: 22701803 Free PMC article. - Use of preclinical models to assess the therapeutic potential of new drug candidates for bladder cancer.
Amit D, Gofrit ON, Matouk I, Birman T, Hochberg A. Amit D, et al. Semin Oncol. 2012 Oct;39(5):534-42. doi: 10.1053/j.seminoncol.2012.08.006. Semin Oncol. 2012. PMID: 23040250 Review. - Long non-coding RNA H19, a novel therapeutic target for pancreatic cancer.
Wang J, Zhao L, Shang K, Liu F, Che J, Li H, Cao B. Wang J, et al. Mol Med. 2020 Apr 9;26(1):30. doi: 10.1186/s10020-020-00156-4. Mol Med. 2020. PMID: 32272875 Free PMC article. Review.
Cited by
- Non-coding RNAs in pancreatic cancer: challenges and opportunities for clinical application.
Taucher V, Mangge H, Haybaeck J. Taucher V, et al. Cell Oncol (Dordr). 2016 Aug;39(4):295-318. doi: 10.1007/s13402-016-0275-7. Epub 2016 Apr 8. Cell Oncol (Dordr). 2016. PMID: 27060060 Review. - Molecular targeted approaches for treatment of pancreatic cancer.
Huang ZQ, Saluja AK, Dudeja V, Vickers SM, Buchsbaum DJ. Huang ZQ, et al. Curr Pharm Des. 2011;17(21):2221-38. doi: 10.2174/138161211796957427. Curr Pharm Des. 2011. PMID: 21777178 Free PMC article. Review. - Noncoding RNAs in cancer and cancer stem cells.
Huang T, Alvarez A, Hu B, Cheng SY. Huang T, et al. Chin J Cancer. 2013 Nov;32(11):582-93. doi: 10.5732/cjc.013.10170. Chin J Cancer. 2013. PMID: 24206916 Free PMC article. Review. - Correlation of long non-coding RNA expression with metastasis, drug resistance and clinical outcome in cancer.
Malek E, Jagannathan S, Driscoll JJ. Malek E, et al. Oncotarget. 2014 Sep 30;5(18):8027-38. doi: 10.18632/oncotarget.2469. Oncotarget. 2014. PMID: 25275300 Free PMC article. Review. - Gene Therapy for Liver Cancers: Current Status from Basic to Clinics.
Kamimura K, Yokoo T, Abe H, Terai S. Kamimura K, et al. Cancers (Basel). 2019 Nov 25;11(12):1865. doi: 10.3390/cancers11121865. Cancers (Basel). 2019. PMID: 31769427 Free PMC article. Review.
References
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. Ca-A Cancer Journal for Clinicians. 2007;57(1):43–66. - PubMed
- Lowenfels AB, Maisonneuve P. Epidemiology and risk factors for pancreatic cancer. Best Practice and Research: Clinical Gastroenterology. 2006;20(2):197–209. - PubMed
- Rosewicz S, Wiedenmann B. Pancreatic carcinoma. The Lancet. 1997;349(9050):485–489. - PubMed
- Sanchez SE, Trevino JG. Current adjuvant and targeted therapies for pancreatic adenocarcinoma. Current Medicinal Chemistry. 2008;15(17):1674–1683. - PubMed
- Ohana P, Bibi O, Matouk I, et al. The use of H19 regulatory sequences for targeted gene therapy in cancer. International Journal of Cancer. 2002;98(5, article 69):645–650. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources