Atomic-level characterization of the ensemble of the Aβ(1-42) monomer in water using unbiased molecular dynamics simulations and spectral algorithms - PubMed (original) (raw)
Atomic-level characterization of the ensemble of the Aβ(1-42) monomer in water using unbiased molecular dynamics simulations and spectral algorithms
Nikolaos G Sgourakis et al. J Mol Biol. 2011.
Abstract
Aβ(1-42) is the highly pathologic isoform of amyloid-β, the peptide constituent of fibrils and neurotoxic oligomers involved in Alzheimer's disease. Recent studies on the structural features of Aβ in water have suggested that the system can be described as an ensemble of distinct conformational species in fast exchange. Here, we use replica exchange molecular dynamics (REMD) simulations to characterize the conformations accessible to Aβ42 in explicit water solvent, under the ff99SB force field. Monitoring the correlation between J-coupling((3)J(H(N))(H(α))) and residual dipolar coupling (RDC) data calculated from the REMD trajectories to their experimental values, as determined by NMR, indicates that the simulations converge towards sampling an ensemble that is representative of the experimental data after 60 ns/replica of simulation time. We further validate the converged MD-derived ensemble through direct comparison with (3)J(H(N))(H(α)) and RDC experimental data. Our analysis indicates that the ff99SB-derived REMD ensemble can reproduce the experimental J-coupling values with high accuracy and further provide good agreement with the RDC data. Our results indicate that the peptide is sampling a highly diverse range of conformations: by implementing statistical learning techniques (Laplacian eigenmaps, spectral clustering, and Laplacian scores), we are able to obtain an otherwise hidden structure in the complex conformational space of the peptide. Using these methods, we characterize the peptide conformations and extract their intrinsic characteristics, identify a small number of different conformations that characterize the whole ensemble, and identify a small number of protein interactions (such as contacts between the peptide termini) that are the most discriminative of the different conformations and thus can be used in designing experimental probes of transitions between such molecular states. This is a study of an important intrinsically disordered peptide system that provides an atomic-level description of structural features and interactions that are relevant during the early stages of the oligomerization and fibril nucleation pathways.
Copyright © 2010. Published by Elsevier Ltd.
Figures
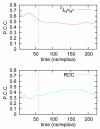
Figure 1. Simulation convergence through comparison with experimental data
Correlation between J-couplings and RDCs calculated from the MD ensemble with their experimentally determined values. Monitoring the Pearson’s correlation coefficient (P.C.C) to the experimental data as a function of the total simulation time indicates that the ensemble is converged after approximately 60ns /replica. The dashed line indicates the start of the production phase of the simulation, during which the calculated J-coupling and RDC values are converged to their ensemble-averaged values, as dictated by the details of the forcefield and solvation model used.
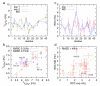
Figure 2. Validation with experimental data
32 experimental three-bond J-coupling values and 22 Residual Dipolar Couplings that report on the average conformation of the backbone and orientation of the amide bond vectors in the molecular alignment frame respectively are compared with results calculated from our REMD simulation trajectories. Two independent experimental measurements of JHNHα3 were used ; . Results are shown in a, c along the sequence of Aβ and as correlation plots in b, d. Experimental J-coupling and RDC values were measured at 300K and 277.3K respectively while simulation values were calculated over the range of replica temperatures 280-310K. Simulation errors were estimated using block averages .
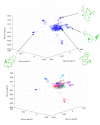
Figure 3. Spectral clustering of the conformational ensemble and identification of representative conformational species
(a): Visualization of the REMD ensemble of conformations in a space defined by the last 3 non-trivial eigenvectors of the Affinity matrix. Good dispersion of the data in this low-dimensional space is observed. Each region of the space contains conformations with distinct contact patterns, as shown in the structural diagrams belonging to individual conformations. (b): Example of the use of the k-means spectral clustering algorithm for k=20, operating in the space defined by the last 6 non-trivial eigenvectors of the Affinity matrix (see methods). The clustering results are visualized in 3 dimensions corresponding to the last 3 non-trivial eigenvectors, same as in (a). Using this technique we have identified several clusters of conformations that share common structural elements as discrete groups in the low-dimensional eigenspace. Representative (central) conformations for selected clusters are shown in the insets, and discussed in the main text. These results suggest that Aβ42 can sample a wide range of conformations with distinct features that can be analyzed using a relatively small set of collective variables.
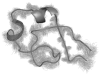
Figure 4. Structural precision in the clustering results
Overlay of all conformations within a single identified cluster according to the spectral clustering technique implemented here. Despite the high degree of structural heterogeneity in the REMD ensemble the contact map-based approach chosen here successfully identifies clusters of conformations with similar features. The central conformation of this cluster is shown in a cartoon representation, while the trace of the backbone is shown for every other member of the cluster. A high degree of structural similarity among conformations within the same cluster is observed, as indicated by an average pairwise RMSD of 1.33Å for the protein backbone among cluster members.
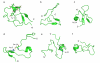
Figure 5. Conformations of Aβ in water
Representative conformations of Aβ42 from the REMD ensemble as identified using the spectral clustering technique. A diverse mixture of extended as well as collapsed coil conformations with secondary structural elements is observed.
Figure 6. Flow diagram of the spectral clustering method
In (a) a diverse ensemble of conformations obtained from enhanced-sampling molecular dynamics is encoded as a binary distance matrix (contact matrix) where each column represents the state of a residue contact (i,j) defined according to a distance threshold of 4.5Å between any pair of heavy atoms belonging to residues i and j. In (b) the original MD dataset, in the contact matrix representation is used to calculate a square Affinity matrix, whose elements are given by e_−_D ij where Dij is the distance between conformations with indices i and j according to a chosen distance kernel. The singular value decomposition of the Affinity matrix yields eigenvectors of high discriminative power. In particular, the m lowest non-trivial eigenvectors (where m<<N) can be used as explicit coordinates to separate the MD ensemble into k clusters using the k-means clustering algorithm.
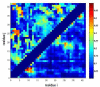
Figure 7. Identification of discriminative contacts using Laplacian scores
The raw probabilities of contact formation between all pairs of residues i,j in Aβ42 according to a 4.5Å distance threshold (lower right quadrant) are contrasted to the extracted Laplacian scores for the same residue pair (upper left quadrant). According to this analysis we identify several contacts of high power in discriminating between different conformational species that are not apparent from a simple inspection of the ensemble-derived statistics of contact formation, as discussed in the text. This information can be used to design experimental probes to investigate transitions between different conformations.
Similar articles
- Characterization of the internal dynamics and conformational space of zinc-bound amyloid β peptides by replica-exchange molecular dynamics simulations.
Xu L, Wang X, Wang X. Xu L, et al. Eur Biophys J. 2013 Jul;42(7):575-86. doi: 10.1007/s00249-013-0906-0. Epub 2013 May 3. Eur Biophys J. 2013. PMID: 23640306 - Aβ monomers transiently sample oligomer and fibril-like configurations: ensemble characterization using a combined MD/NMR approach.
Rosenman DJ, Connors CR, Chen W, Wang C, García AE. Rosenman DJ, et al. J Mol Biol. 2013 Sep 23;425(18):3338-59. doi: 10.1016/j.jmb.2013.06.021. Epub 2013 Jun 25. J Mol Biol. 2013. PMID: 23811057 Free PMC article. - Effects of force fields on the conformational and dynamic properties of amyloid β(1-40) dimer explored by replica exchange molecular dynamics simulations.
Watts CR, Gregory A, Frisbie C, Lovas S. Watts CR, et al. Proteins. 2018 Mar;86(3):279-300. doi: 10.1002/prot.25439. Epub 2017 Dec 25. Proteins. 2018. PMID: 29235155 - Understanding amyloid fibril nucleation and aβ oligomer/drug interactions from computer simulations.
Nguyen P, Derreumaux P. Nguyen P, et al. Acc Chem Res. 2014 Feb 18;47(2):603-11. doi: 10.1021/ar4002075. Epub 2013 Dec 24. Acc Chem Res. 2014. PMID: 24368046 Review. - Molecular Dynamics Simulations Combined with Nuclear Magnetic Resonance and/or Small-Angle X-ray Scattering Data for Characterizing Intrinsically Disordered Protein Conformational Ensembles.
Chan-Yao-Chong M, Durand D, Ha-Duong T. Chan-Yao-Chong M, et al. J Chem Inf Model. 2019 May 28;59(5):1743-1758. doi: 10.1021/acs.jcim.8b00928. Epub 2019 Mar 18. J Chem Inf Model. 2019. PMID: 30840442 Review.
Cited by
- Promotion and Inhibition of Amyloid-β Peptide Aggregation: Molecular Dynamics Studies.
Itoh SG, Okumura H. Itoh SG, et al. Int J Mol Sci. 2021 Feb 13;22(4):1859. doi: 10.3390/ijms22041859. Int J Mol Sci. 2021. PMID: 33668406 Free PMC article. Review. - Reliable oligonucleotide conformational ensemble generation in explicit solvent for force field assessment using reservoir replica exchange molecular dynamics simulations.
Henriksen NM, Roe DR, Cheatham TE 3rd. Henriksen NM, et al. J Phys Chem B. 2013 Apr 18;117(15):4014-27. doi: 10.1021/jp400530e. Epub 2013 Apr 4. J Phys Chem B. 2013. PMID: 23477537 Free PMC article. - Pharmacological affinity fingerprints derived from bioactivity data for the identification of designer drugs.
He K. He K. J Cheminform. 2022 Jun 7;14(1):35. doi: 10.1186/s13321-022-00607-6. J Cheminform. 2022. PMID: 35672835 Free PMC article. - Characterization of the internal dynamics and conformational space of zinc-bound amyloid β peptides by replica-exchange molecular dynamics simulations.
Xu L, Wang X, Wang X. Xu L, et al. Eur Biophys J. 2013 Jul;42(7):575-86. doi: 10.1007/s00249-013-0906-0. Epub 2013 May 3. Eur Biophys J. 2013. PMID: 23640306 - Side-chain dynamics reveals transient association of Aβ(1-40) monomers with amyloid fibers.
Krishnamoorthy J, Brender JR, Vivekanandan S, Jahr N, Ramamoorthy A. Krishnamoorthy J, et al. J Phys Chem B. 2012 Nov 26;116(46):13618-23. doi: 10.1021/jp305279w. Epub 2012 Nov 12. J Phys Chem B. 2012. PMID: 23116141 Free PMC article.
References
- Soto C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat Rev Neurosci. 2003;4:49–60. - PubMed
- Rauk A. The chemistry of Alzheimer’s disease. Chemical Society Reviews. 2009;38:2698–2715. - PubMed
- Lesne S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. A specific amyloid-beta protein assembly in the brain impairs memory. Nature. 2006;440:352–7. - PubMed
- Mucke L, Masliah E, Yu GQ, Mallory M, Rockenstein EM, Tatsuno G, Hu K, Kholodenko D, Johnson-Wood K, McConlogue L. High-level neuronal expression of abeta 1-42 in wild-type human amyloid protein precursor transgenic mice: synaptotoxicity without plaque formation. J Neurosci. 2000;20:4050–8. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous