Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells - PubMed (original) (raw)
. 2010 Dec;42(12):1113-7.
doi: 10.1038/ng.710. Epub 2010 Nov 7.
Moran N Cabili, Mitchell Guttman, Yuin-Han Loh, Kelly Thomas, In Hyun Park, Manuel Garber, Matthew Curran, Tamer Onder, Suneet Agarwal, Philip D Manos, Sumon Datta, Eric S Lander, Thorsten M Schlaeger, George Q Daley, John L Rinn
Affiliations
- PMID: 21057500
- PMCID: PMC3040650
- DOI: 10.1038/ng.710
Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells
Sabine Loewer et al. Nat Genet. 2010 Dec.
Erratum in
- Nat Genet. 2010 Dec;42(12): 3 p following 1117
Abstract
The conversion of lineage-committed cells to induced pluripotent stem cells (iPSCs) by reprogramming is accompanied by a global remodeling of the epigenome, resulting in altered patterns of gene expression. Here we characterize the transcriptional reorganization of large intergenic non-coding RNAs (lincRNAs) that occurs upon derivation of human iPSCs and identify numerous lincRNAs whose expression is linked to pluripotency. Among these, we defined ten lincRNAs whose expression was elevated in iPSCs compared with embryonic stem cells, suggesting that their activation may promote the emergence of iPSCs. Supporting this, our results indicate that these lincRNAs are direct targets of key pluripotency transcription factors. Using loss-of-function and gain-of-function approaches, we found that one such lincRNA (lincRNA-RoR) modulates reprogramming, thus providing a first demonstration for critical functions of lincRNAs in the derivation of pluripotent stem cells.
Figures
Figure 1
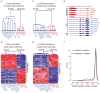
Direct reprogramming of fibroblasts converts both protein-coding gene and lincRNA expression to a puripotent cell-specific profile. A, C) Unsupervised hierarchical clustering of protein-coding gene expression (A) and lincRNA expression (C) segregates fibroblasts (red) from ESCs and fibroblast-derived iPSCs (blue). B, D) Supervised hierarchical clustering analysis identifies 6865 protein-coding genes (B) and 237 lincRNAs (D) that are differentially expressed between ESCs/iPSCs and fibroblasts (genes: >2fold, P<0.05; lincRNAs: >2fold, FWER<0.05). Expression values are represented in shades of red and blue relative to being above (red) or below (blue) the median expression value across all samples (log scale 2, from -3 to +3). hFib2 fibroblasts are represented as two replicates (hFib2 and hFib2a). E) Examples of reprogrammed lincRNAs; left: lincRNA expressed in all fibroblasts is repressed in all pluripotent cells, right: a pluripotent cell-specific lincRNA that becomes activated during reprogramming. Expression values for each tiled probe (x-axis) are displayed as normalized hybridization intensity (y-axis). F) Correlation analysis of lincRNAs and neighboring genes. Density plot of multiple testing corrected p-values (x-axis) for lincRNAs that are positively (blue) or negatively (red) correlated with their protein-coding gene neighbors.
Figure 2
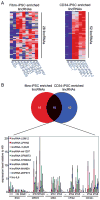
Several lincRNAs show enriched expression in iPSCs compared with ESCs. A) Heatmap of 28 and 52 lincRNAs that are more highly expressed in fibroblast-derived iPSCs (left) and CD34+-derived iPSCs (right), respectively, compared with ESCs (>2fold, FWER<0.05). Expression values are represented in shades of red and blue relative to being above (red) or below (blue) the median expression value across all samples (log scale 2, from -3 to +3). B) Top: Venn Diagram shows 10 lincRNAs that are commonly enriched in fibroblast- and CD34+-derived iPSCs. Bottom: qRT-PCR validation of the 10 commonly enriched lincRNAs (named according to their 3’ protein-coding gene neighbor) across three human ESC lines (H1, H9, BG01), fibroblasts (MRC5, MSC, hFib2) and CD34+ cells, and their derivative iPSC lines. Expression values are represented relative to the RNA levels in H9 ESCs.
Figure 3
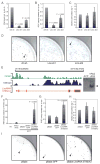
Transcriptional regulation of iPSC-enriched lincRNAs. A) iPSC-enriched lincRNA loci are bound by pluripotency transcription factors. Top: lincRNA loci demarcated by domains enriched in histone H3K4me3 indicating RNA polymerase II promoters and H3K36me3 indicating regions of transcriptional elongation, in human ESCs (green and blue, respectively). Bottom: ChIP in hFib2-iPS5 cells followed by quantitative PCR analysis detects binding of OCT4, SOX2, and NANOG within lincRNA-SFMBT2, lincRNA-VLDLR, and lincRNA-ST8SIA3 regions close to lincRNA promoter regions (peaks of H3K4me). ChIP enrichment values are displayed normalized to a control region (chr12: 7,839,777- 7,839,966; hg18); anti-GFP ChIP was used as a negative control. Positions of ChIP-PCR fragments are indicated by black lines. B) Changes in iPSC-enriched lincRNA levels upon siRNA-mediated knock-down of OCT4 in iPSC. Top: qRT-PCR of OCT4, NANOG and LMNA transcript levels upon depletion of OCT4. Bottom: qRT-PCR of iPSC-enriched lincRNA levels upon depletion of OCT4. Transcript levels are displayed relative to non-targeting control siRNAs (ctrl siRNA) (n=3, error bars +/-s.e.m). C) iPSC-enriched lincRNA expression during EB differentiation. Top: qRT-PCR analysis monitoring transcript levels of pluripotency markers (OCT4 and NANOG) and the differentiation marker LMNA over a ten day differentiation time-course; bottom: qRT-PCR analysis of iPSC-enriched lincRNAs. RNA levels are depicted relative to undifferentiated cells on day 0 (n=3, error bars +/-s.e.m).
Figure 4
LincRNA-ST8SIA3 expression modulates reprogramming. A) qRT-PCR verifies lincRNA-ST8SIA3 knock-down with Linc-sh1 and Linc-sh2 in hFib2-iPS5 cells relative to a non-targeting shRNA control (n=2, error bar: +/-s.e.m). B) Quantification of Tra-1-60+ iPSC colonies upon knock-down of lincRNA-ST8SIA3 relative to the control (day 21; n=4, error bar: +/-s.e.m). C) Quantification of cell numbers on day 6-7 of reprogramming in lincRNA-ST8SIA3 shRNA samples relative to the control (n=4, error bar: +/-s.e.m). D) Images showing quarters of Tra-1-60 stained reprogramming plates upon infection of a non-targeting control and two lincRNA-ST8SIA3 targeting shRNAs. Arrowheads mark Tra-1-60+ iPSC colonies. E) Structure of the lincRNA-ST8SIA3 locus. Green, Blue: Demarcation of the H3K4me-H3K36me domain in ESCs. Red: Structure of lincRNA-ST8SIA3 RNA; asterisk marks position of OCT4/SOX2/NANOG binding (see Figure 3A). Right: Northern hybridization of lincRNA-ST8SIA3 detects a 2.6kb transcript in hFib2-iSP5, but not dH1f (full-length blot see Supplementary Figure 12). F) qRT-PCR verifies lincRNA-ST8SIA3 overexpression from a retroviral vector (pBabe-lincRNA-ST8SIA3) compared with pBabe-puro and pBabe-puro-GFP vectors in dH1f relative to the levels in H9 ESCs and hFib2-iPS5 (n=2, error bars: +/-s.e.m). G) Quantification of Tra-1-60+ iPSC colonies upon overexpression of lincRNA-ST8SIA3 compared to pBabe and pBabe-GFP controls (n=5, error bar: +/-s.e.m.). H) Quantification of cell numbers on day 6-7 in lincRNA-ST8SIA3 overexpressing cells and controls. Cell numbers are relative to the pBabe control (day 28 +/-2; n=5, error bar: +/- s.e.m.). I) Image of quarter-plates of Tra-1-60 stained colonies (arrowheads) in pBabe, pBabe-GFP, and pBabe-lincRNA-ST8SIA3 infected samples. Statistical analysis was performed using Student’s t-test.
Comment in
- LincRNAs join the pluripotency alliance.
Ng JH, Ng HH. Ng JH, et al. Nat Genet. 2010 Dec;42(12):1035-6. doi: 10.1038/ng1210-1035. Nat Genet. 2010. PMID: 21102618 No abstract available.
Similar articles
- lincRNAs act in the circuitry controlling pluripotency and differentiation.
Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. Guttman M, et al. Nature. 2011 Aug 28;477(7364):295-300. doi: 10.1038/nature10398. Nature. 2011. PMID: 21874018 Free PMC article. - Long noncoding RNAs sustain high expression levels of exogenous octamer-binding protein 4 by sponging regulatory microRNAs during cellular reprogramming.
Zhang X, Zhang J, Zheng K, Zhang H, Pei X, Yin Z, Wen D, Kong Q. Zhang X, et al. J Biol Chem. 2019 Nov 22;294(47):17863-17874. doi: 10.1074/jbc.RA119.010284. Epub 2019 Oct 17. J Biol Chem. 2019. PMID: 31624145 Free PMC article. - LincRNAs join the pluripotency alliance.
Ng JH, Ng HH. Ng JH, et al. Nat Genet. 2010 Dec;42(12):1035-6. doi: 10.1038/ng1210-1035. Nat Genet. 2010. PMID: 21102618 No abstract available. - An Insight into Reprogramming Barriers to iPSC Generation.
Haridhasapavalan KK, Raina K, Dey C, Adhikari P, Thummer RP. Haridhasapavalan KK, et al. Stem Cell Rev Rep. 2020 Feb;16(1):56-81. doi: 10.1007/s12015-019-09931-1. Stem Cell Rev Rep. 2020. PMID: 31758374 Review. - Role of Long Non-coding RNAs in Reprogramming to Induced Pluripotency.
Kanwal S, Guo X, Ward C, Volpe G, Qin B, Esteban MA, Bao X. Kanwal S, et al. Genomics Proteomics Bioinformatics. 2020 Feb;18(1):16-25. doi: 10.1016/j.gpb.2019.06.003. Epub 2020 May 21. Genomics Proteomics Bioinformatics. 2020. PMID: 32445708 Free PMC article. Review.
Cited by
- LncRNA BCAR4 wires up signaling transduction in breast cancer.
Xing Z, Park PK, Lin C, Yang L. Xing Z, et al. RNA Biol. 2015;12(7):681-9. doi: 10.1080/15476286.2015.1053687. RNA Biol. 2015. PMID: 26016563 Free PMC article. - Nothing in Evolution Makes Sense Except in the Light of Genomics: Read-Write Genome Evolution as an Active Biological Process.
Shapiro JA. Shapiro JA. Biology (Basel). 2016 Jun 8;5(2):27. doi: 10.3390/biology5020027. Biology (Basel). 2016. PMID: 27338490 Free PMC article. Review. - Comprehensive analysis of the lncRNA-miRNA-mRNA regulatory network for bladder cancer.
Huang M, Long Y, Jin Y, Ya W, Meng D, Qin T, Su L, Zhou W, Wu J, Huang C, Huang Q. Huang M, et al. Transl Androl Urol. 2021 Mar;10(3):1286-1301. doi: 10.21037/tau-21-81. Transl Androl Urol. 2021. PMID: 33850763 Free PMC article. - Exosomes in disease and regeneration: biological functions, diagnostics, and beneficial effects.
Lin Y, Anderson JD, Rahnama LMA, Gu SV, Knowlton AA. Lin Y, et al. Am J Physiol Heart Circ Physiol. 2020 Dec 1;319(6):H1162-H1180. doi: 10.1152/ajpheart.00075.2020. Epub 2020 Sep 28. Am J Physiol Heart Circ Physiol. 2020. PMID: 32986962 Free PMC article. - Regulatory non-coding RNAs in pluripotent stem cells.
Rosa A, Brivanlou AH. Rosa A, et al. Int J Mol Sci. 2013 Jul 11;14(7):14346-73. doi: 10.3390/ijms140714346. Int J Mol Sci. 2013. PMID: 23852015 Free PMC article. Review.
References
- Maherali N, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K08 HL089150/HL/NHLBI NIH HHS/United States
- DP2 OD006670/OD/NIH HHS/United States
- 1DP2OD00667-01/OD/NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- RC2 HL102815/HL/NHLBI NIH HHS/United States
- DP2 OD006670-01/OD/NIH HHS/United States
- 1 RC2-HL102815/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials