CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes - PubMed (original) (raw)
CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes
Vikas Rishi et al. Proc Natl Acad Sci U S A. 2010.
Abstract
DNA methylation of the cytosine in the CpG dinucleotide is typically associated with gene silencing. Genomic analyses have identified low CpG promoters that are both methylated and transcriptionally active, but the mechanism underlying the activation of these methylated promoters remains unclear. Here we show that CpG methylation of the CRE sequence (TGACGTCA) enhances the DNA binding of the C/EBPα transcription factor, a protein critical for activation of differentiation in various cell types. Transfection assays also show that C/EBPα activates the CRE sequence only when it is methylated. The biological significance of this observation was seen in differentiating primary keratinocyte cultures from newborn mice where certain methylated promoters are both bound by C/EBPα and activated upon differentiation. Experimental demethylation by either 5-azacytidine treatment or DNMT1 depletion diminished both C/EBPα binding and activation of the same methylated promoters upon differentiation suggesting that CpG methylation can localize C/EBPα. Transfection studies in cell cultures using methylated tissue-specific proximal promoters identified half-CRE (CGTCA) and half-C/EBP (CGCAA) sequences that need to be methylated for C/EBPα mediated activation. In primary dermal fibroblasts, C/EBPα activates a different set of methylated tissue-specific promoters upon differentiation into adipocytes. These data identify a new function for methyl CpGs: producing DNA binding sites at half-CRE and half-C/EBP sequences for C/EBPα that are needed to activate tissue-specific genes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
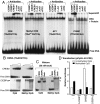
Fig. 1.
C/EBP family members bind to methylated consensus CRE site. (A) EMSA with primary keratinocyte nuclear extracts using antibodies to B-ZIP proteins and 28 bp oligonucleotides containing (i) an unmethylated CRE, (ii) a methylated CRE, (iii) an AP-1 consensus site, or (iv) a C/EBP consensus site identifies that CREB binds to an unmethylated CRE sequence, whereas C/EBP family members bind to a methylated CRE sequence. Only c-Jun and JunD bind the AP-1 sequence, and only C/EBP family members bind the C/EBP sequence. (B) EMSA using the CRE sequence used in A and increasing concentrations of pure CREB and C/EBPα B-ZIP protein domains (5-, 15-, 50-, and 150-nM dimer) show CREB preferentially binds the unmethylated CRE, whereas C/EBPα preferentially binds the methylated CRE. (C) EMSA showing a mixture of CREB and C/EBPα B-ZIP domains at equimolar concentrations (50-nM dimer). (D) pCpGL-4X-CRE, a luciferase reporter containing four consensus CRE sites, was constructed from pCpGL, a parental luciferase reporter plasmid without CpGs in the backbone. Cotransfection of pCpGL-4X-CRE into keratinocytes with CREB coding plasmid increases the activity of only the unmethylated reporter, whereas C/EBPα coding plasmid increases the activity of only the methylated reporter (T test, p < 0.001).
Fig. 2.
C/EBPα binding activates methylated promoters in keratinocyte differentiation. (A) Scatter plot (log 2) of MeDIP enrichment before and after keratinocyte differentiation using Nimblegen mouse promoter arrays with each dot representing a promoter. (B) MeDIP enrichment in undifferentiated keratinocytes versus the number of CpG dinucleotides in the promoter (-1,000 bp to +500 bp). The red line demarcates two groups of promoters, unmethylated (UM) and methylated (M). (C) Methylation status of promoters for the most abundantly expressed genes (top 20%) in undifferentiated (Undiff) keratinocytes. (D) Number of UM and M promoters whose mRNA expression was doubled with differentiation (Diff, Upper). mRNA induction during differentiation is preferentially inhibited (induction decreases > 50%) for genes with methylated promoters by experimental demethylation using either 5-aza (*_χ_2 test, p < 0.01), or siRNA to DNMT1. Both demethylation methods decreased expression of the same methylated promoters (hypergeometric test, _p_ < 0.05). (_E_) Number of UM and M promoters bound by C/EBPα (binding enrichment > 1.2, T test, p < 0.03) in undifferentiated keratinocytes. Logo showing a C/EBP consensus site is the most enriched sequence in promoters bound by C/EBPα. C/EBPα binding is inhibited (< 0.8) preferentially at methylated promoters after treatment with 5-aza (*_χ_2 test, p < 0.001) or siRNA to DNMT1 (*_χ_2 test, p < 0.02). Both demethylation methods decreased C/EBPα binding to the same methylated promoters (hypergeometric test, p < 0.001). The half-CRE (CGTCA) is the most common DNA sequence occurring in promoters with decreased C/EBPα binding following either demethylation method. (F) qPCR of C/EBPα ChIP DNA from undifferentiated keratinocytes for the tissue-specific (Bglap-rs1 and AK043395) and housekeeping (Rps12) promoters with or without 5-aza treatment compared to input DNA. All values are expressed as fold change normalized to Rps12 (*T test, p < 0.05).
Fig. 3.
C/EBPα activates methylated promoters in keratinocytes. (A) Bisulfite sequencing of the cloned keratinocyte specific Bglap-rs1 proximal promoter before and after differentiation shows that it is predominantly methylated. Filled circles denote methylated CpG and open circles denote unmethylated CpG. Half-C/EBP (-37 bp) (1) and half-CRE (-417 bp) (2) sequences are marked. (B) Luciferase activities of Bglap-rs1 promoter (pCpGL-Bglap-rs1) cotransfected with CREB or C/EBPα coding plasmid show that CREB activates the unmethylated plasmid, whereas C/EBPα activates the methylated plasmid (T test, p < 0.05). Promoters with the mutated half-C/EBP (1) or half-CRE (2) sequence have reduced activity. All reporter activities are expressed relative to the unmethylated plasmid. (C) EMSA showing CREB and C/EBPα (15-, 50-, 150-, and 500-nM dimer) binding to the unmethylated and methylated half-C/EBP (1) or half-CRE (2) sequences from Bglap-rs1. (D) Mixture of CREB and C/EBPα (each 500-nM dimer) shows C/EBPα binds preferentially to the methylated half-CRE sequence even in the presence of equimolar CREB. CREB and C/EBPα cannot bind the mutant half-CRE site of Bglap-rs1 (
SI Appendix, Fig. S4_I_
). (E) Fold difference in C/EBPα binding to methylated (M) compared to unmethylated (UM) DNA sequences determined by EMSA versus fold difference in C/EBPα transactivation of a methylated reporter compared to an unmethylated reporter. Seven sequences are plotted and show a positive correlation; i.e., preferential binding to the methylated sequence produces preferential transactivation of the methylated luciferase reporter.
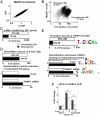
Fig. 4.
C/EBPα binding activates methylated promoters in fibroblasts. (A) Oil-Red-O staining showing accumulation of neutral lipids in newborn dermal fibroblast cultures 8 d after induction for adipogenic differentiation. Differentiation is inhibited by 5-aza treatment, depletion of DNMT1, or expression of A-C/EBP, a dominant negative to C/EBP family members. (B) Number of UM and M promoters whose mRNA expression was doubled with differentiation (Upper). mRNA induction during differentiation is preferentially inhibited (induction decreases > 50%) for genes with methylated promoters by experimental demethylation using either 5-aza (*_χ_2 test, p < 0.05), or siRNA to DNMT1 or A-C/EBP. (_C_) Number of UM and M promoters bound by C/EBPα (binding enrichment > 1.2, T test, p < 0.03) in adipocytes. Logo showing a C/EBPα consensus site is the most enriched sequence in these promoters. (Lower) Decreased C/EBPα binding (< 0.8) after demethylation preferentially at methylated promoters by 5-aza (* _χ_2 test, p < 0.05) or siRNA to DNMT1. Both demethylation methods decreased C/EBPα binding to the same methylated promoters (hypergeometric test, p < 0.02). Logos containing a CpG are the most common DNA sequences occurring in promoters with decreased C/EBPα binding following either demethylation method. (D) Bisulfite sequencing of the adipocyte specific promoter, Gpd1, showing it is predominantly methylated before and after differentiation. Filled circles denote methylated CpG and open circles denote unmethylated CpG. The half-CRE sequence is marked by an arrow. Activity of a luciferase reporter containing the Gpd1 promoter (pCpGL-Gpd1), either unmethylated or enzymatically methylated, transiently transfected into differentiated fibroblasts. Cotransfection with expression plasmids encoding CREB or C/EBPα enhanced activity of unmethylated and methylated promoters, respectively (T test, p < 0.05). Mutation of half-CRE site reduced activity of the promoter. EMSA showing CREB and C/EBPα (15-, 50-, 150-, and 500-nM dimer) binding to the unmethylated and methylated half-CRE sequence from Gpd1 promoter. Mixture of CREB and C/EBPα (150-nM dimer each) shows C/EBPα binds preferentially to the methylated sequence even in the presence of equimolar CREB.
Similar articles
- Epigenetic modulation of tumor suppressor CCAAT/enhancer binding protein alpha activity in lung cancer.
Tada Y, Brena RM, Hackanson B, Morrison C, Otterson GA, Plass C. Tada Y, et al. J Natl Cancer Inst. 2006 Mar 15;98(6):396-406. doi: 10.1093/jnci/djj093. J Natl Cancer Inst. 2006. PMID: 16537832 - All and only CpG containing sequences are enriched in promoters abundantly bound by RNA polymerase II in multiple tissues.
Rozenberg JM, Shlyakhtenko A, Glass K, Rishi V, Myakishev MV, FitzGerald PC, Vinson C. Rozenberg JM, et al. BMC Genomics. 2008 Feb 5;9:67. doi: 10.1186/1471-2164-9-67. BMC Genomics. 2008. PMID: 18252004 Free PMC article. - CG methylation.
Vinson C, Chatterjee R. Vinson C, et al. Epigenomics. 2012 Dec;4(6):655-63. doi: 10.2217/epi.12.55. Epigenomics. 2012. PMID: 23244310 Free PMC article. Review. - Sequence determinants, function, and evolution of CpG islands.
Angeloni A, Bogdanovic O. Angeloni A, et al. Biochem Soc Trans. 2021 Jun 30;49(3):1109-1119. doi: 10.1042/BST20200695. Biochem Soc Trans. 2021. PMID: 34156435 Free PMC article. Review.
Cited by
- Maternal deprivation in rats is associated with corticotrophin-releasing hormone (CRH) promoter hypomethylation and enhances CRH transcriptional responses to stress in adulthood.
Chen J, Evans AN, Liu Y, Honda M, Saavedra JM, Aguilera G. Chen J, et al. J Neuroendocrinol. 2012 Jul;24(7):1055-64. doi: 10.1111/j.1365-2826.2012.02306.x. J Neuroendocrinol. 2012. PMID: 22375940 Free PMC article. - Preferential CEBP binding to T:G mismatches and increased C-to-T human somatic mutations.
Yang J, Horton JR, Akdemir KC, Li J, Huang Y, Kumar J, Blumenthal RM, Zhang X, Cheng X. Yang J, et al. Nucleic Acids Res. 2021 May 21;49(9):5084-5094. doi: 10.1093/nar/gkab276. Nucleic Acids Res. 2021. PMID: 33877329 Free PMC article. - Epigenetic control in skin development, homeostasis and injury repair.
Kang S, Chovatiya G, Tumbar T. Kang S, et al. Exp Dermatol. 2019 Apr;28(4):453-463. doi: 10.1111/exd.13872. Epub 2019 Feb 12. Exp Dermatol. 2019. PMID: 30624812 Free PMC article. Review. - Silencing of CEBPB-AS1 modulates CEBPB expression and resensitizes BRAF-inhibitor resistant melanoma cells to vemurafenib.
Vidarsdottir L, Fernandes RV, Zachariadis V, Das I, Edsbäcker E, Sigvaldadottir I, Azimi A, Höiom V, Hansson J, Grandér D, Egyházi Brage S, Pokrovskaja Tamm K. Vidarsdottir L, et al. Melanoma Res. 2020 Oct;30(5):443-454. doi: 10.1097/CMR.0000000000000675. Melanoma Res. 2020. PMID: 32467529 Free PMC article. - DNA methylation and differentiation: silencing, upregulation and modulation of gene expression.
Ehrlich M, Lacey M. Ehrlich M, et al. Epigenomics. 2013;5(5):553-68. doi: 10.2217/epi.13.43. Epigenomics. 2013. PMID: 24059801 Free PMC article. Review.
References
- Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. - PubMed
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. - PubMed
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources