NBC: the Naive Bayes Classification tool webserver for taxonomic classification of metagenomic reads - PubMed (original) (raw)
NBC: the Naive Bayes Classification tool webserver for taxonomic classification of metagenomic reads
Gail L Rosen et al. Bioinformatics. 2011.
Abstract
Motivation: Datasets from high-throughput sequencing technologies have yielded a vast amount of data about organisms in environmental samples. Yet, it is still a challenge to assess the exact organism content in these samples because the task of taxonomic classification is too computationally complex to annotate all reads in a dataset. An easy-to-use webserver is needed to process these reads. While many methods exist, only a few are publicly available on webservers, and out of those, most do not annotate all reads.
Results: We introduce a webserver that implements the naïve Bayes classifier (NBC) to classify all metagenomic reads to their best taxonomic match. Results indicate that NBC can assign next-generation sequencing reads to their taxonomic classification and can find significant populations of genera that other classifiers may miss.
Availability: Publicly available at: http://nbc.ece.drexel.edu.
Figures
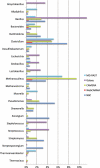
Fig. 1.
Percentage of reads that are assigned to a particular genera out of all 454 reads from the Biogas reactor community. CAMERA and NBC tend to agree for over 70% of the genera shown while MG-RAST agrees with CAMERA and NBC near 50%. WebCARMA bins fewers reads, and Galaxy has high variability. For the first 5602 reads (1.5 Mb web site limit), Phylopythia only classifies eight reads to the phylum level and is not included in the graph due to its inability to make assignments at the genus level.
Similar articles
- NBC update: The addition of viral and fungal databases to the Naïve Bayes classification tool.
Rosen GL, Lim TY. Rosen GL, et al. BMC Res Notes. 2012 Jan 31;5:81. doi: 10.1186/1756-0500-5-81. BMC Res Notes. 2012. PMID: 22293603 Free PMC article. - MNBC: a multithreaded Minimizer-based Naïve Bayes Classifier for improved metagenomic sequence classification.
Lu R, Dumonceaux T, Anzar M, Zovoilis A, Antonation K, Barker D, Corbett C, Nadon C, Robertson J, Eagle SHC, Lung O, Rudar J, Surujballi O, Laing C. Lu R, et al. Bioinformatics. 2024 Oct 1;40(10):btae601. doi: 10.1093/bioinformatics/btae601. Bioinformatics. 2024. PMID: 39388213 Free PMC article. - The Naïve Bayes classifier++ for metagenomic taxonomic classification-query evaluation.
Duan HN, Hearne G, Polikar R, Rosen GL. Duan HN, et al. Bioinformatics. 2024 Dec 26;41(1):btae743. doi: 10.1093/bioinformatics/btae743. Bioinformatics. 2024. PMID: 39700412 Free PMC article. - AmphoraNet: the webserver implementation of the AMPHORA2 metagenomic workflow suite.
Kerepesi C, Bánky D, Grolmusz V. Kerepesi C, et al. Gene. 2014 Jan 10;533(2):538-40. doi: 10.1016/j.gene.2013.10.015. Epub 2013 Oct 19. Gene. 2014. PMID: 24144838 - Metagenomic search strategies for interactions among plants and multiple microbes.
Melcher U, Verma R, Schneider WL. Melcher U, et al. Front Plant Sci. 2014 Jun 11;5:268. doi: 10.3389/fpls.2014.00268. eCollection 2014. Front Plant Sci. 2014. PMID: 24966863 Free PMC article. Review.
Cited by
- Computational meta'omics for microbial community studies.
Segata N, Boernigen D, Tickle TL, Morgan XC, Garrett WS, Huttenhower C. Segata N, et al. Mol Syst Biol. 2013 May 14;9:666. doi: 10.1038/msb.2013.22. Mol Syst Biol. 2013. PMID: 23670539 Free PMC article. Review. - Overview of Trends in the Application of Metagenomic Techniques in the Analysis of Human Enteric Viral Diversity in Africa's Environmental Regimes.
Osunmakinde CO, Selvarajan R, Sibanda T, Mamba BB, Msagati TAM. Osunmakinde CO, et al. Viruses. 2018 Aug 14;10(8):429. doi: 10.3390/v10080429. Viruses. 2018. PMID: 30110939 Free PMC article. Review. - Overview of Virus Metagenomic Classification Methods and Their Biological Applications.
Nooij S, Schmitz D, Vennema H, Kroneman A, Koopmans MPG. Nooij S, et al. Front Microbiol. 2018 Apr 23;9:749. doi: 10.3389/fmicb.2018.00749. eCollection 2018. Front Microbiol. 2018. PMID: 29740407 Free PMC article. Review. - A Bio Medical Waste Identification and Classification Algorithm Using Mltrp and Rvm.
Achuthan A, Ayyallu Madangopal V. Achuthan A, et al. Iran J Public Health. 2016 Oct;45(10):1276-1287. Iran J Public Health. 2016. PMID: 27957434 Free PMC article. - High-throughput sequencing of 16S rDNA amplicons characterizes bacterial composition in bronchoalveolar lavage fluid in patients with ventilator-associated pneumonia.
Yang XJ, Wang YB, Zhou ZW, Wang GW, Wang XH, Liu QF, Zhou SF, Wang ZH. Yang XJ, et al. Drug Des Devel Ther. 2015 Aug 18;9:4883-96. doi: 10.2147/DDDT.S87634. eCollection 2015. Drug Des Devel Ther. 2015. PMID: 26345636 Free PMC article.
References
- Altschul SF, et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
- Hery M, et al. Monitoring of bacterial communities during low temperature thermal treatment of activated sludge combining dna phylochip and respirometry techniques. Water Res. 2010 [Epub ahead of print, doi: 10.1016/j.watres.2010.07.003.] - PubMed
- McHardy AC, et al. Accurate phylogenetic classification of variable-length dna fragments. Nat. Methods. 2007;4:63–72. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources