Reactome: a database of reactions, pathways and biological processes - PubMed (original) (raw)
. 2011 Jan;39(Database issue):D691-7.
doi: 10.1093/nar/gkq1018. Epub 2010 Nov 9.
Gavin O'Kelly, Guanming Wu, Robin Haw, Marc Gillespie, Lisa Matthews, Michael Caudy, Phani Garapati, Gopal Gopinath, Bijay Jassal, Steven Jupe, Irina Kalatskaya, Shahana Mahajan, Bruce May, Nelson Ndegwa, Esther Schmidt, Veronica Shamovsky, Christina Yung, Ewan Birney, Henning Hermjakob, Peter D'Eustachio, Lincoln Stein
Affiliations
- PMID: 21067998
- PMCID: PMC3013646
- DOI: 10.1093/nar/gkq1018
Reactome: a database of reactions, pathways and biological processes
David Croft et al. Nucleic Acids Res. 2011 Jan.
Abstract
Reactome (http://www.reactome.org) is a collaboration among groups at the Ontario Institute for Cancer Research, Cold Spring Harbor Laboratory, New York University School of Medicine and The European Bioinformatics Institute, to develop an open source curated bioinformatics database of human pathways and reactions. Recently, we developed a new web site with improved tools for pathway browsing and data analysis. The Pathway Browser is an Systems Biology Graphical Notation (SBGN)-based visualization system that supports zooming, scrolling and event highlighting. It exploits PSIQUIC web services to overlay our curated pathways with molecular interaction data from the Reactome Functional Interaction Network and external interaction databases such as IntAct, BioGRID, ChEMBL, iRefIndex, MINT and STRING. Our Pathway and Expression Analysis tools enable ID mapping, pathway assignment and overrepresentation analysis of user-supplied data sets. To support pathway annotation and analysis in other species, we continue to make orthology-based inferences of pathways in non-human species, applying Ensembl Compara to identify orthologs of curated human proteins in each of 20 other species. The resulting inferred pathway sets can be browsed and analyzed with our Species Comparison tool. Collaborations are also underway to create manually curated data sets on the Reactome framework for chicken, Drosophila and rice.
Figures
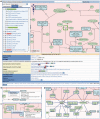
Figure 1.
The Reactome pathway browser and the molecular overlays. (A) The main features of the pathway browser are the ‘Search’ bar at the top, the ‘Pathways’ panel on the left, the ‘Visualization’ panel on the right and a ‘Details’ panel (hidden by default when the pathway browser first loads). When a pathway is selected from the ‘Pathways’ panel hierarchy, it is highlighted in bright green in the hierarchy and its parent terms are highlighted in yellow–green. A description of any reaction or pathway in the pathway diagrams can be displayed below the diagram in the ‘Details’ panel by selecting the event within the diagram. Right-clicking the mouse while the cursor is over a physical entity displays the context sensitive menus. (B) Selecting ‘Other Pathways’ displays additional Reactome pathways that contain the highlighted physical entity. Navigating to the additional pathway is achieved by clicking the pathway name. (C) Selecting ‘Participating Molecules’ displays additional components of the macromolecular complex. Displaying the component information in the ‘Details’ panel is achieved by clicking the entity name. (D) Protein–protein interactions are displayed in the pathway browser for SHC1 [cytosol] and SHP2 [cytosol] physical entities.
Figure 2.
Species comparison tool and model organism pathway diagrams. (A) A drop-down menu is used to select the model organism species. (B) Results for the comparison of human and mouse pathways. Each row in the table is a pathway; the columns are pathway name, model organism name, number of proteins in the human pathway, number of orthologous proteins in the inferred model organism pathway, a graphical representation of the ratio of these two values and a ‘View’ button that creates a pathway diagram. ‘Sort’ buttons at the top of each column allow the table to be re-ordered according to cell contents in that column. (C) The pathway browser displaying the comparison of the human and mouse Mitotic M-M/G1 pathways. Physical entities in the pathway diagram are color-coded: gray, no match; black, a complex (multicomponent) entity; and yellow, the protein’s ortholog is present in human. (D) The pathway browser displaying the inferred mouse Mitotic M-M/G1 pathway.
Figure 3.
The pathway and expression analysis tools. (A) The results table for the ‘ID mapping and pathway assignment’. The sortable table contains one row for each Reactome pathway and four additional columns: repeats the user-supplied IDs, the corresponding UniProt ID, the species name and names of pathways in which this ID can be found. The names/IDs in the last two columns are clickable links that take the user to a diagram of the named pathway. (B) The results for the ‘overrepresentation analysis’, presented as a list of clickable links of enriched events. The warmer the color, the higher the level of overrepresentation in the given pathway. Clicking on the ‘+’ next to the pathway name gives access to the user-supplied identifiers that are found in the pathway, along with the corresponding UniProt IDs. (C) The results table for the expression analysis. The sortable table contains one row for each Reactome pathway and five additional columns: name of the pathway, Species of presented results, total number of proteins in pathway, number of proteins in the user-supplied data that fall into the pathway, graphical representation of the ratio of these two values and a ‘View’ button that creates a pathway diagram. (D) The pathway browser displaying the colored physical entities that correspond to expression values of the experimental data. The nodes in this diagram have a special color-coding: gray, no match; black, a (multicomponent) complex entity; and other colors represent expression levels. If the numerical data are a time series, the grey bar at the bottom of the colored pathway diagram allows the user to step through time points and visualize changes in expression levels with time of the individual genes involved in the pathway.
Similar articles
- Reactome pathway analysis to enrich biological discovery in proteomics data sets.
Haw R, Hermjakob H, D'Eustachio P, Stein L. Haw R, et al. Proteomics. 2011 Sep;11(18):3598-613. doi: 10.1002/pmic.201100066. Proteomics. 2011. PMID: 21751369 Free PMC article. - Using the reactome database.
Haw R, Stein L. Haw R, et al. Curr Protoc Bioinformatics. 2012 Jun;Chapter 8:8.7.1-8.7.23. doi: 10.1002/0471250953.bi0807s38. Curr Protoc Bioinformatics. 2012. PMID: 22700314 Free PMC article. - Using the Reactome Database.
Rothfels K, Milacic M, Matthews L, Haw R, Sevilla C, Gillespie M, Stephan R, Gong C, Ragueneau E, May B, Shamovsky V, Wright A, Weiser J, Beavers D, Conley P, Tiwari K, Jassal B, Griss J, Senff-Ribeiro A, Brunson T, Petryszak R, Hermjakob H, D'Eustachio P, Wu G, Stein L. Rothfels K, et al. Curr Protoc. 2023 Apr;3(4):e722. doi: 10.1002/cpz1.722. Curr Protoc. 2023. PMID: 37053306 Free PMC article. - Plant Reactome and PubChem: The Plant Pathway and (Bio)Chemical Entity Knowledgebases.
Gupta P, Naithani S, Preece J, Kim S, Cheng T, D'Eustachio P, Elser J, Bolton EE, Jaiswal P. Gupta P, et al. Methods Mol Biol. 2022;2443:511-525. doi: 10.1007/978-1-0716-2067-0_27. Methods Mol Biol. 2022. PMID: 35037224 Review. - A survey of metabolic databases emphasizing the MetaCyc family.
Karp PD, Caspi R. Karp PD, et al. Arch Toxicol. 2011 Sep;85(9):1015-33. doi: 10.1007/s00204-011-0705-2. Epub 2011 Apr 27. Arch Toxicol. 2011. PMID: 21523460 Free PMC article. Review.
Cited by
- A Computational Protocol for the Knowledge-Based Assessment and Capture of Pathologies.
Page J, Moore N, Broderick G. Page J, et al. Methods Mol Biol. 2025;2868:265-284. doi: 10.1007/978-1-0716-4200-9_14. Methods Mol Biol. 2025. PMID: 39546235 - Knowledge graph embeddings in the biomedical domain: are they useful? A look at link prediction, rule learning, and downstream polypharmacy tasks.
Gema AP, Grabarczyk D, De Wulf W, Borole P, Alfaro JA, Minervini P, Vergari A, Rajan A. Gema AP, et al. Bioinform Adv. 2024 Jul 17;4(1):vbae097. doi: 10.1093/bioadv/vbae097. eCollection 2024. Bioinform Adv. 2024. PMID: 39506988 Free PMC article. Review. - Human DCP1 is crucial for mRNA decapping and possesses paralog-specific gene regulating functions.
Chen TW, Liao HW, Noble M, Siao JY, Cheng YH, Chiang WC, Lo YT, Chang CT. Chen TW, et al. Elife. 2024 Nov 1;13:RP94811. doi: 10.7554/eLife.94811. Elife. 2024. PMID: 39485278 Free PMC article. - BACH1 as a key driver in rheumatoid arthritis fibroblast-like synoviocytes identified through gene network analysis.
Pelissier A, Laragione T, Harris C, Rodríguez Martínez M, Gulko PS. Pelissier A, et al. Life Sci Alliance. 2024 Oct 28;8(1):e202402808. doi: 10.26508/lsa.202402808. Print 2025 Jan. Life Sci Alliance. 2024. PMID: 39467637 Free PMC article. - Construction and validation of cell cycle-related prognostic genetic model for glioblastoma.
Zhou R, Zhang K, Dai T, Guo Z, Li T, Hong X. Zhou R, et al. Medicine (Baltimore). 2024 Oct 4;103(40):e39205. doi: 10.1097/MD.0000000000039205. Medicine (Baltimore). 2024. PMID: 39465756 Free PMC article.
References
- Degtyarenko K, Hastings J, de Matos P, Ennis M. ChEBI: an open bioinformatics and cheminformatics resource. Curr. Protoc. Bioinform. 2009 Chapter 14: Unit 4.9. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources