Analysis of codon usage in bovine viral diarrhea virus - PubMed (original) (raw)
Analysis of codon usage in bovine viral diarrhea virus
Meng Wang et al. Arch Virol. 2011 Jan.
Abstract
Bovine viral diarrhea virus (BVDV) is a widespread virus in beef and dairy herds. BVDV has been grouped into two genotypes, genotype 1 and genotype 2. In this study, the relative synonymous codon usage (RSCU) values, effective number of codon (ENC) values and nucleotide content were investigated, and a comparative analysis of codon usage patterns for open reading frames (ORFs) of 22 BVDV genomes, including 14 of genotype 1 and 8 of genotype 2, was carried out. A high A+U content and low codon bias were found in BVDV genomes. Depending on the RSCU data, it was found that there was a significant variation in bias of codon usage between the two genotypes, and a geographic factor exists only in genotype-1 of BVDV. The RSCU data have a negative correlation with general average hydrophobicity (GRAVY), aromaticity and nucleotide content. Furthermore, the overall abundance of C and U has no effect on the synonymous codon usage patterns. In contrast, the A and G content showed a significant correlation with the nucleotide content at the third position. In addition, the codon usage patterns of BVDV are similar to those of 22 conserved genes of Bos taurus. Taken together, the genetic characteristics of BVDV possibly result from interactions between natural selection and mutation pressure.
Figures
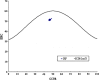
Fig. 1
The relationship between the effective number of codons (ENC) and the GC content of the third codon position (GC3)
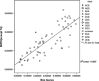
Fig. 2
A plot of average proportions of codons of BVDV within its synonymous family (excluding strain no. 14, which was isolated from swine) and Bos taurus (excluding the codons for Trp, Met and the three termination codons)
Similar articles
- Analysis of codon usage and nucleotide composition bias in polioviruses.
Zhang J, Wang M, Liu WQ, Zhou JH, Chen HT, Ma LN, Ding YZ, Gu YX, Liu YS. Zhang J, et al. Virol J. 2011 Mar 30;8:146. doi: 10.1186/1743-422X-8-146. Virol J. 2011. PMID: 21450075 Free PMC article. - Patterns and influencing factor of synonymous codon usage in porcine circovirus.
Liu XS, Zhang YG, Fang YZ, Wang YL. Liu XS, et al. Virol J. 2012 Mar 15;9:68. doi: 10.1186/1743-422X-9-68. Virol J. 2012. PMID: 22416942 Free PMC article. - Comparative characterization analysis of synonymous codon usage bias in classical swine fever virus.
Xu X, Fei D, Han H, Liu H, Zhang J, Zhou Y, Xu C, Wang H, Cao H, Zhang H. Xu X, et al. Microb Pathog. 2017 Jun;107:368-371. doi: 10.1016/j.micpath.2017.04.019. Epub 2017 Apr 14. Microb Pathog. 2017. PMID: 28416383 - The characteristic of codon usage pattern and its evolution of hepatitis C virus.
Hu JS, Wang QQ, Zhang J, Chen HT, Xu ZW, Zhu L, Ding YZ, Ma LN, Xu K, Gu YX, Liu YS. Hu JS, et al. Infect Genet Evol. 2011 Dec;11(8):2098-102. doi: 10.1016/j.meegid.2011.08.025. Epub 2011 Sep 1. Infect Genet Evol. 2011. PMID: 21907310 - Variability and Global Distribution of Subgenotypes of Bovine Viral Diarrhea Virus.
Yeşilbağ K, Alpay G, Becher P. Yeşilbağ K, et al. Viruses. 2017 May 26;9(6):128. doi: 10.3390/v9060128. Viruses. 2017. PMID: 28587150 Free PMC article. Review.
Cited by
- Analysis of codon usage and nucleotide composition bias in polioviruses.
Zhang J, Wang M, Liu WQ, Zhou JH, Chen HT, Ma LN, Ding YZ, Gu YX, Liu YS. Zhang J, et al. Virol J. 2011 Mar 30;8:146. doi: 10.1186/1743-422X-8-146. Virol J. 2011. PMID: 21450075 Free PMC article. - Phylogenetic and codon usage analysis of atypical porcine pestivirus (APPV).
Pan S, Mou C, Wu H, Chen Z. Pan S, et al. Virulence. 2020 Dec;11(1):916-926. doi: 10.1080/21505594.2020.1790282. Virulence. 2020. PMID: 32615860 Free PMC article. - Analysis of synonymous codon usage patterns in duck hepatitis A virus: a comparison on the roles of mutual pressure and natural selection.
Chen Y, Chen YF. Chen Y, et al. Virusdisease. 2014;25(3):285-93. doi: 10.1007/s13337-014-0191-2. Epub 2014 Jan 25. Virusdisease. 2014. PMID: 25674595 Free PMC article. - Patterns and influencing factor of synonymous codon usage in porcine circovirus.
Liu XS, Zhang YG, Fang YZ, Wang YL. Liu XS, et al. Virol J. 2012 Mar 15;9:68. doi: 10.1186/1743-422X-9-68. Virol J. 2012. PMID: 22416942 Free PMC article.
References
- Adams MJ, Antoniw JF. Codon usage bias amongst plant viruses. Arch Virol. 2004;149:113–135. - PubMed
- Castillo-Davis CI, Hartl DL. Genome evolution and developmental constraint in Caenorhabditis elegans. Mol Biol Evol. 2002;19:728–735. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources