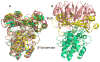Protein binding specificity versus promiscuity - PubMed (original) (raw)
Review
Protein binding specificity versus promiscuity
Gideon Schreiber et al. Curr Opin Struct Biol. 2011 Feb.
Abstract
Interactions between macromolecules in general, and between proteins in particular, are essential for any life process. Examples include transfer of information, inhibition or activation of function, molecular recognition as in the immune system, assembly of macromolecular structures and molecular machines, and more. Proteins interact with affinities ranging from millimolar to femtomolar and, because affinity determines the concentration required to obtain 50% binding, the amount of different complexes formed is very much related to local concentrations. Although the concentration of a specific binding partner is usually quite low in the cell (nanomolar to micromolar), the total concentration of other macromolecules is very high, allowing weak and non-specific interactions to play important roles. In this review we address the question of binding specificity, that is, how do some proteins maintain monogamous relations while others are clearly polygamous. We examine recent work that addresses the molecular and structural basis for specificity versus promiscuity. We show through examples how multiple solutions exist to achieve binding via similar interfaces and how protein specificity can be tuned using both positive and negative selection (specificity by demand). Binding of a protein to numerous partners can be promoted through variation in which residues are used for binding, conformational plasticity and/or post-translational modification. Natively unstructured regions represent the extreme case in which structure is obtained only upon binding. Many natively unstructured proteins serve as hubs in protein-protein interaction networks and such promiscuity can be of functional importance in biology.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures
Figure 1
Overlay of β-lactamase – BLIP structures. A. cyan – TEM1-BLIP (1JTG), magenta – SHV-BLIP (2G2U), yellow – KPC2-BLIP (3E2L), pink – TEM1-BLIP1 (3GMW) and green – is BLP (3GMX) overlaid on TEM1-BLIP. B. TEM1-BLIP2 (1JTD) structure (pink over cyan) overlaid on TEM1-BLIP.
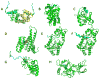
Figure 2
Interactions formed by p53 residues 367–391. In each panel, the p53 fragment is in cyan. The p53 sequence is 367-SHLKSKKGQSTSRHKKLMFKTEGPD-391, and in each structure the bold lysine residues are shown in stick representation, if unmodified. Post-translationally modified residues are shown using spheres. (A) S100 calcium-binding protein with p53 residues 367–388 (1DT7) [72], (B) Cyclin A2 with p53 residues 378–386 (1H26) [73], (C) Tumor suppressor p53-binding protein 1 with p53 dimethylated lysine residue 382 (3LGL); residues 377–381 and 383–387 did not show clear density [59], (D) 14-3-3 protein sigma with p53 residues 385–391, phosphothreonine 387 (3LW1) [60], (E) NAD-dependent deacetylase Sir2 with p53 residues 378–384 (2H2F) [74], (F) NAD-dependent deacetylase Sir2 with p53 residues 373–385; acetyllysine 382 (2H2D) [74], (G) CREB-binding protein with p53 residues 367–386; acetyllysine 382 (1JSP) [75], (H) Histone-lysine N-methyltransferase with p53 residues 369–374; N-methyl-lysine 372 (1XQH) [76].
Similar articles
- Protein promiscuity: drug resistance and native functions--HIV-1 case.
Fernández A, Tawfik DS, Berkhout B, Sanders R, Kloczkowski A, Sen T, Jernigan B. Fernández A, et al. J Biomol Struct Dyn. 2005 Jun;22(6):615-24. doi: 10.1080/07391102.2005.10531228. J Biomol Struct Dyn. 2005. PMID: 15842167 - Intrinsically unstructured proteins and neurodegenerative diseases: conformational promiscuity at its best.
Das S, Mukhopadhyay D. Das S, et al. IUBMB Life. 2011 Jul;63(7):478-88. doi: 10.1002/iub.498. IUBMB Life. 2011. PMID: 21698751 Review. - Structural characterisation and functional significance of transient protein-protein interactions.
Nooren IM, Thornton JM. Nooren IM, et al. J Mol Biol. 2003 Jan 31;325(5):991-1018. doi: 10.1016/s0022-2836(02)01281-0. J Mol Biol. 2003. PMID: 12527304 - Evolution of specificity in protein-protein interactions.
Peleg O, Choi JM, Shakhnovich EI. Peleg O, et al. Biophys J. 2014 Oct 7;107(7):1686-96. doi: 10.1016/j.bpj.2014.08.004. Biophys J. 2014. PMID: 25296322 Free PMC article. - Flexible nets. The roles of intrinsic disorder in protein interaction networks.
Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Dunker AK, et al. FEBS J. 2005 Oct;272(20):5129-48. doi: 10.1111/j.1742-4658.2005.04948.x. FEBS J. 2005. PMID: 16218947 Review.
Cited by
- The coming of age of de novo protein design.
Huang PS, Boyken SE, Baker D. Huang PS, et al. Nature. 2016 Sep 15;537(7620):320-7. doi: 10.1038/nature19946. Nature. 2016. PMID: 27629638 Review. - A Critical Analysis of the FDA's Omics-Driven Pharmacodynamic Biomarkers to Establish Biosimilarity.
Niazi SK. Niazi SK. Pharmaceuticals (Basel). 2023 Nov 2;16(11):1556. doi: 10.3390/ph16111556. Pharmaceuticals (Basel). 2023. PMID: 38004421 Free PMC article. Review. - Designing specific protein-protein interactions using computation, experimental library screening, or integrated methods.
Chen TS, Keating AE. Chen TS, et al. Protein Sci. 2012 Jul;21(7):949-63. doi: 10.1002/pro.2096. Epub 2012 Jun 8. Protein Sci. 2012. PMID: 22593041 Free PMC article. Review. - Characterization of 14-3-3-ζ Interactions with integrin tails.
Bonet R, Vakonakis I, Campbell ID. Bonet R, et al. J Mol Biol. 2013 Sep 9;425(17):3060-72. doi: 10.1016/j.jmb.2013.05.024. Epub 2013 Jun 11. J Mol Biol. 2013. PMID: 23763993 Free PMC article. - Structural Basis for Multi-specificity of MRG Domains.
Xie T, Zmyslowski AM, Zhang Y, Radhakrishnan I. Xie T, et al. Structure. 2015 Jun 2;23(6):1049-57. doi: 10.1016/j.str.2015.03.020. Epub 2015 May 7. Structure. 2015. PMID: 25960410 Free PMC article.
References
- Wang Y, Li C, Pielak GJ. Effects of proteins on protein diffusion. J Am Chem Soc. 2010;132:9392–9397. In this NMR study it is shown that weak interactions at high concentrations in an E.coli cell lysate significantly affect the rotational diffusion of a protein, an effect that is absent when using synthetic polymers, which apparently are inert to the probed protein. - PMC - PubMed
- Greenspan NS. Cohen’s Conjecture, Howard’s Hypothesis, and Ptashne’s Ptruth: an exploration of the relationship between affinity and specificity. Trends Immunol. 2010;31:138–143. - PubMed
- Neuvirth H, Raz R, Schreiber G. ProMate: A Structure Based Prediction Program to Identify the Location of Protein-Protein Binding Sites. J Mol Biol. 2004;338:181–199. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM067681/GM/NIGMS NIH HHS/United States
- R01 GM067681-07/GM/NIGMS NIH HHS/United States
- R01 GM084181/GM/NIGMS NIH HHS/United States
- R01 GM084181-03/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources