Does M. tuberculosis genomic diversity explain disease diversity? - PubMed (original) (raw)
Does M. tuberculosis genomic diversity explain disease diversity?
Mireilla Coscolla et al. Drug Discov Today Dis Mech. 2010 Spring.
Abstract
The outcome of tuberculosis infection and disease is highly variable. This variation has been attributed primarily to host and environmental factors, but better understanding of the global genomic diversity in the M. tuberculosis complex (MTBC) suggests that bacterial factors could also be involved. Review of nearly 100 published reports shows that MTBC strains differ in their virulence and immunogenicity in experimental models, but whether this phenotypic variation plays a role in human disease remains unclear. Given the complex interactions between the host, the pathogen and the environment, linking MTBC genotypic diversity to experimental and clinical phenotypes requires an integrated systems epidemiology approach embedded in a robust evolutionary framework.
Figures
Figure 1. Neighbour-joining phylogeny based on 9,037 variable common nucleotide positions across 21 human MTBC genome sequences
Six main lineages are defined within human-adapted MTBC. The six lineages correspond to groups previously detected with a variety of genotyping techniques. Asterisks indicate spoligotype patterns within the main lineages which are considered phylogenetically unrelated based on spoligotyping, but are in fact related based on LSP analysis and genome sequencing [108] (adapted from Ref. 38). Abbreviations: MLSA=Multi Locus Sequence Analysis; LSP=Long Sequence Polymorphisms; SNP= Single nucleotide polymorphism; PGG=Principal Genetic Group.
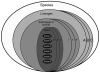
Figure 2. Theoretical approach defining a set of hierarchical strain groupings within a given bacterial species
Our capability to define different genetic entities as cluster, family, sub-lineage or lineage depends on the breadth of our sampling and molecular markers used. In this case, the species could be divided in different lineages, and within each lineage in different sub-lineages, clusters or families. The lower grouping is the strain or clone, which has been defined as a set of stocks that share many identical or similar properties due to a recent common clonal ancestry [109]. Here a clone represents genetically identical or almost identical bacteria, and can include different isolates from one patient, a transmission chain or any related units which share a very recent common ancestor.
Similar articles
- Biological and Epidemiological Consequences of MTBC Diversity.
Coscolla M. Coscolla M. Adv Exp Med Biol. 2017;1019:95-116. doi: 10.1007/978-3-319-64371-7_5. Adv Exp Med Biol. 2017. PMID: 29116631 Review. - The Bioinformatics Analysis of Comparative Genomics of Mycobacterium tuberculosis Complex (MTBC) Provides Insight into Dissimilarities between Intraspecific Groups Differing in Host Association, Virulence, and Epitope Diversity.
Jia X, Yang L, Dong M, Chen S, Lv L, Cao D, Fu J, Yang T, Zhang J, Zhang X, Shang Y, Wang G, Sheng Y, Huang H, Chen F. Jia X, et al. Front Cell Infect Microbiol. 2017 Mar 21;7:88. doi: 10.3389/fcimb.2017.00088. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28377903 Free PMC article. - Shaping the niche in macrophages: Genetic diversity of the M. tuberculosis complex and its consequences for the infected host.
Reiling N, Homolka S, Kohl TA, Steinhäuser C, Kolbe K, Schütze S, Brandenburg J. Reiling N, et al. Int J Med Microbiol. 2018 Jan;308(1):118-128. doi: 10.1016/j.ijmm.2017.09.009. Epub 2017 Sep 14. Int J Med Microbiol. 2018. PMID: 28969988 Review. - Genetic diversity in Mycobacterium tuberculosis.
Gagneux S. Gagneux S. Curr Top Microbiol Immunol. 2013;374:1-25. doi: 10.1007/82_2013_329. Curr Top Microbiol Immunol. 2013. PMID: 23677208 - Human tuberculosis and Mycobacterium tuberculosis complex: A review on genetic diversity, pathogenesis and omics approaches in host biomarkers discovery.
Kanabalan RD, Lee LJ, Lee TY, Chong PP, Hassan L, Ismail R, Chin VK. Kanabalan RD, et al. Microbiol Res. 2021 May;246:126674. doi: 10.1016/j.micres.2020.126674. Epub 2021 Jan 29. Microbiol Res. 2021. PMID: 33549960 Review.
Cited by
- Plasma Level of IL-4 Differs in Patients Infected with Different Modern Lineages of M. tuberculosis.
Mihret A, Bekele Y, Loxton AG, Aseffa A, Howe R, Walzl G. Mihret A, et al. J Trop Med. 2012;2012:518564. doi: 10.1155/2012/518564. Epub 2012 Sep 25. J Trop Med. 2012. PMID: 23049571 Free PMC article. - Molecular characterisation of multidrug-resistant Mycobacterium tuberculosis isolates from a high-burden tuberculosis state in Brazil.
Salvato RS, Schiefelbein S, Barcellos RB, Praetzel BM, Anusca IS, Esteves LS, Halon ML, Unis G, Dias CF, Miranda SS, de Almeida IN, de Assis Figueredo LJ, Silva EC, Kritski AL, Dalla Costa ER, Rossetti MLR. Salvato RS, et al. Epidemiol Infect. 2019 Jan;147:e216. doi: 10.1017/S0950268819001006. Epidemiol Infect. 2019. PMID: 31364547 Free PMC article. - Isotuberculosinol: the unusual case of an immunomodulatory diterpenoid from Mycobacterium tuberculosis.
Mann FM, Peters RJ. Mann FM, et al. Medchemcomm. 2012 Aug 1;3(8):899-904. doi: 10.1039/c2md20030a. Medchemcomm. 2012. PMID: 23926455 Free PMC article. No abstract available. - "Pseudo-Beijing": evidence for convergent evolution in the direct repeat region of Mycobacterium tuberculosis.
Fenner L, Malla B, Ninet B, Dubuis O, Stucki D, Borrell S, Huna T, Bodmer T, Egger M, Gagneux S. Fenner L, et al. PLoS One. 2011;6(9):e24737. doi: 10.1371/journal.pone.0024737. Epub 2011 Sep 13. PLoS One. 2011. PMID: 21935448 Free PMC article.
References
- Loo IHM, et al. Temporal trends in the population structure of Bordetella pertussis during 1949-1996 in a highly vaccinated population. J Infect Dis. 1999;179:915–923. - PubMed
LinkOut - more resources
Full Text Sources