The spliceosomal proteome: at the heart of the largest cellular ribonucleoprotein machine - PubMed (original) (raw)
Review
The spliceosomal proteome: at the heart of the largest cellular ribonucleoprotein machine
Saba Valadkhan et al. Proteomics. 2010 Nov.
Abstract
Almost all primary transcripts in higher eukaryotes undergo several splicing events and alternative splicing is a major factor in generating proteomic diversity. Thus, the spliceosome, the ribonucleoprotein assembly that performs splicing, is a highly critical cellular machine and as expected, a very complex one. Indeed, the spliceosome is one of the largest, if not the largest, molecular machine in the cell with over 150 different components in human. A large fraction of the spliceosomal proteome is organized into small nuclear ribonucleoprotein particles by associating with one of the small nuclear RNAs, and the function of many spliceosomal proteins revolve around their association or interaction with the spliceosomal RNAs or the substrate pre-messenger RNAs. In addition to the complex web of protein-RNA interactions, an equally complex network of protein-protein interactions exists in the spliceosome, which includes a number of large, conserved proteins with critical functions in the spliceosomal catalytic core. These include the largest conserved nuclear protein, Prp8, which plays a critical role in spliceosomal function in a hitherto unknown manner. Taken together, the large spliceosomal proteome and its dynamic nature has made it a highly challenging system to study, and at the same time, provides an exciting example of the evolution of a proteome around a backbone of primordial RNAs likely dating from the RNA World.
Copyright © 2010 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
Conflict of interest statement
The author declares no conflict of interest.
Figures
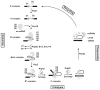
Fig. 1. Schematic representation of a group II self-splicing intron, from which spliceosomes may have evolved
The exons are shown in gray rectangles. The identity of each RNA domain is indicated close to each domain. The sites of 5′ and 3′ splice sites and branch site are marked as 5′SS, 3′SS and BS, respectively. The functional and/or structural homologue of each RNA element in the spliceosome is shown in gray circles.
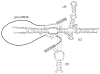
Fig. 2. The interaction of U6, U5 and U2 snRNAs with the pre-mRNA before the first step of splicing
The basepairing interactions within each snRNA, between U6 and U2 snRNAs and between snRNAs and the pre-mRNA are shown. Numbers reflect the human numbering system. The exons in pre-mRNA are shown as solid rectangles, with the intron shown as a line. The sequence of the 5′ splice site and branch site are shown.
Fig. 3. The RNA and protein composition of each snRNP particle
Note that all snRNPs except U6 carry a set of Sm proteins, whereas U6 instead contains LSm proteins.
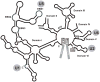
Fig. 4. The spliceosomal cycle
The pre-mRNA is shown in black, and the location of the branch site is marked by an A. The exons are shown as solid rectangles and the intron is drawn as a thin line. The spliceosomal complexes and snRNPs are shown in gray. The identity of each spliceosomal complex and snRNP particle is shown. The main steps of spliceosomal cycle, assembly, catalysis, disassembly and recycling are marked on the graph. The main proofreading steps of spliceosomal cycle are marked by clock signs and the main RNA helicases involved in each conformational rearrangement are indicated. The snRNAs within each spliceosomal complex are shown.
Similar articles
- Structural bioinformatics of the human spliceosomal proteome.
Korneta I, Magnus M, Bujnicki JM. Korneta I, et al. Nucleic Acids Res. 2012 Aug;40(15):7046-65. doi: 10.1093/nar/gks347. Epub 2012 May 9. Nucleic Acids Res. 2012. PMID: 22573172 Free PMC article. - Comprehensive proteomic analysis of the human spliceosome.
Zhou Z, Licklider LJ, Gygi SP, Reed R. Zhou Z, et al. Nature. 2002 Sep 12;419(6903):182-5. doi: 10.1038/nature01031. Nature. 2002. PMID: 12226669 - Structural dynamics of the N-terminal domain and the Switch loop of Prp8 during spliceosome assembly and activation.
Jia X, Sun C. Jia X, et al. Nucleic Acids Res. 2018 May 4;46(8):3833-3840. doi: 10.1093/nar/gky242. Nucleic Acids Res. 2018. PMID: 29635373 Free PMC article. Review. - The spliceosome: the most complex macromolecular machine in the cell?
Nilsen TW. Nilsen TW. Bioessays. 2003 Dec;25(12):1147-9. doi: 10.1002/bies.10394. Bioessays. 2003. PMID: 14635248 Review. - Spliceosomal proteomics in Trypanosoma brucei reveal new RNA splicing factors.
Luz Ambrósio D, Lee JH, Panigrahi AK, Nguyen TN, Cicarelli RM, Günzl A. Luz Ambrósio D, et al. Eukaryot Cell. 2009 Jul;8(7):990-1000. doi: 10.1128/EC.00075-09. Epub 2009 May 8. Eukaryot Cell. 2009. PMID: 19429779 Free PMC article.
Cited by
- The RNA polymerase C-terminal domain: a new role in spliceosome assembly.
David CJ, Manley JL. David CJ, et al. Transcription. 2011 Sep-Oct;2(5):221-5. doi: 10.4161/trns.2.5.17272. Transcription. 2011. PMID: 22231118 Free PMC article. - Breast Cancer and the Other Non-Coding RNAs.
Dvorská D, Braný D, Ňachajová M, Halašová E, Danková Z. Dvorská D, et al. Int J Mol Sci. 2021 Mar 23;22(6):3280. doi: 10.3390/ijms22063280. Int J Mol Sci. 2021. PMID: 33807045 Free PMC article. Review. - Gene ontology analysis of expanded porcine blastocysts from gilts fed organic or inorganic selenium combined with pyridoxine.
Dalto DB, Tsoi S, Dyck MK, Matte JJ. Dalto DB, et al. BMC Genomics. 2018 Nov 21;19(1):836. doi: 10.1186/s12864-018-5237-1. BMC Genomics. 2018. PMID: 30463510 Free PMC article. - Functional Analysis of Mutations in Exon 9 of NF1 Reveals the Presence of Several Elements Regulating Splicing.
Hernández-Imaz E, Martín Y, de Conti L, Melean G, Valero A, Baralle M, Hernández-Chico C. Hernández-Imaz E, et al. PLoS One. 2015 Oct 28;10(10):e0141735. doi: 10.1371/journal.pone.0141735. eCollection 2015. PLoS One. 2015. PMID: 26509978 Free PMC article. - Regulation of chemoresistance via alternative messenger RNA splicing.
Eblen ST. Eblen ST. Biochem Pharmacol. 2012 Apr 15;83(8):1063-72. doi: 10.1016/j.bcp.2011.12.041. Epub 2012 Jan 8. Biochem Pharmacol. 2012. PMID: 22248731 Free PMC article. Review.
References
- Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. - PubMed
- Newman AJ, Nagai K. Structural studies of the spliceosome: blind men and an elephant. Curr Opin Struct Biol. 2010;20:82–89. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous