RioK1, a new interactor of protein arginine methyltransferase 5 (PRMT5), competes with pICln for binding and modulates PRMT5 complex composition and substrate specificity - PubMed (original) (raw)
RioK1, a new interactor of protein arginine methyltransferase 5 (PRMT5), competes with pICln for binding and modulates PRMT5 complex composition and substrate specificity
Gernot Guderian et al. J Biol Chem. 2011.
Abstract
Protein arginine methylation plays a critical role in differential gene expression through modulating protein-protein and protein-DNA/RNA interactions. Although numerous proteins undergo arginine methylation, only limited information is available on how protein arginine methyltransferases (PRMTs) identify their substrates. The human PRMT5 complex consists of PRMT5, WD45/MEP50 (WD repeat domain 45/methylosome protein 50), and pICln and catalyzes the symmetrical arginine dimethylation of its substrate proteins. pICln recruits the spliceosomal Sm proteins to the PRMT5 complex for methylation, which allows their subsequent loading onto snRNA to form small nuclear ribonucleoproteins. To understand how the PRMT5 complex is regulated, we investigated its biochemical composition and identified RioK1 as a novel, stoichiometric component of the PRMT5 complex. We show that RioK1 and pICln bind to PRMT5 in a mutually exclusive fashion. This results in a PRMT5-WD45/MEP50 core structure that either associates with pICln or RioK1 in distinct complexes. Furthermore, we show that RioK1 functions in analogy to pICln as an adapter protein by recruiting the RNA-binding protein nucleolin to the PRMT5 complex for its symmetrical methylation. The exclusive interaction of PRMT5 with either pICln or RioK1 thus provides the first mechanistic insight into how a methyltransferase can distinguish between its substrate proteins.
Figures
FIGURE 1.
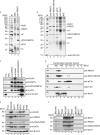
RioK1 is a novel stoichiometric component of the human PRMT5 complex. A, the PRMT5 complex was immunoprecipitated from HeLa extract using anti-PRMT5 antibodies covalently coupled to protein A-Sepharose, resolved via SDS-PAGE, and visualized by silver staining. Normal rabbit serum (NRS) coupled to protein A-Sepharose was used as control (lane 2). The _asterisk_-marked band at ∼90 kDa was excised and identified as RioK1 by mass spectrometry (lane 3). HC and LC represent the heavy and light chain of co-eluted antibodies. Lane 1 shows protein standard (M). B, the PRMT5 complex was immunoprecipitated from HeLa extract using antibodies directed against PRMT5 (lane 2), WD45/MEP50 (lane 3), pICln (lane 4), RioK1 (lane 5), or normal rabbit serum (NRS) as control (lane 6). The precipitated complex components were analyzed by SDS-PAGE and silver staining. Note that protein content was normalized to PRMT5, explaining the different intensities of co-precipitated pICln in A (lane 3) and B (lane 2). C, the PRMT5 complex was immunoprecipitated similarly to B, resolved by SDS-PAGE, and analyzed by Western blotting using the indicated antibodies. D, HeLa total cell lysate (TCL) was separated by gel filtration chromatography, and fractions were analyzed by Western blotting. E, total protein extracts were generated from MCF7 (breast cancer cell line), HeLa (cervix carcinoma cell line), Kelly (neuroblastoma cell line), NEC8 (testicular tumor cell line), HEK293T (embryonic kidney cell line), MelHo (melanoma cell line), and HS68 (primary foreskin fibroblast cell line) and normalized to β-actin (lowest panel). Expression of RioK1 and the PRMT5 complex components PRMT5, WD45/MEP50, and pICln were analyzed by immunoblotting with specific antibodies as indicated. F, to analyze whether RioK1 is part of the PRMT5 complex in different cell lines, RioK1 was immunoprecipitated from MCF7, HEK293T, and HS68 extracts. Co-precipitation of PRMT5 complex components was assessed by Western blotting with specific antibodies as indicated (lanes 3–5). Normal rabbit serum was used as control (lanes 6–8).
FIGURE 2.
RioK1 is an exclusively cytoplasmic protein. A, HeLa cells were grown on coverslips, fixed, permeabilized, and stained with antibodies against PRMT5, WD45/MEP50, pICln, and RioK1. DAPI was used to visualize DNA in the nucleus (upper panel). Cytoplasmic localization of RioK1 was confirmed by merge of DAPI and rhodamin channels. B, GFP-RioK1 was overexpressed in HeLa cells for 48 h and compared with GFP expression for control (lower panel). C, HeLa cells were fractionated into total cell lysate (TCL, lane 1), cytoplasmic extract (CPE, lane 2), and nuclear extract (NE, lane 3). The fractions were analyzed by Western blotting using the indicated antibodies.
FIGURE 3.
RioK1 directly interacts with PRMT5 via its very N-terminal region. A, in an interaction assay, in vitro translated, radioactively [35S]methionine-labeled RioK1 was incubated with GST-tagged PRMT5 (lane 2), WD45/MEP50 (lane 3), pICln (lane 4), and GST (lane 5) bound to GSH-Sepharose. After extensive washing, the precipitated proteins were eluted with sample buffer, resolved by SDS-PAGE, and visualized by Coomassie staining. The in vitro translated protein was visualized by autoradiography of the dried gel. For comparison, 10% of in vitro translated RioK1 was loaded (lane 6). B, GST-tagged truncations of RioK1 comprising aa 1–242 (lanes 2–4), aa 227–568 (lanes 5–7), aa 1–120 (lanes 8–10), or GST (lanes 11–13) bound to GSH-Sepharose were incubated with in vitro translated, [35S]methionine-labeled full-length (fl), N-terminal (nt; aa 1–291), or C-terminal (ct; aa 295–637) versions of PRMT5 in an interaction assay. For comparison, 10% of in vitro translated proteins were loaded (lanes 14–16). The interaction assay was analyzed as described in A. C, GST-PRMT5 (lanes 2–6) and GST (lanes 7–11) bound to GSH-Sepharose were incubated with an in vitro translated, [35S]methionine-labeled version of RioK, including full-length RioK1, aa 1–242, aa 227–568, aa 1–120, and aa 121–242. For comparison, 10% of in vitro translated RioK1 was loaded (lanes 12–16). D, GST-tagged pICln (lanes 2 and 3), RioK1 (lanes 4–6), GST alone (lanes 7 and 8), RioK2 (lanes 9 and 10), or RioK3 (lanes 11 and 12), covalently cross-linked to GSH-Sepharose were incubated with or without HeLa extract in a pulldown assay. As a control, non-cross-linked protein was analyzed by SDS-PAGE. After extensive washing, the precipitated proteins were eluted from the matrix by sample buffer, resolved by SDS-PAGE, visualized via Coomassie staining, and analyzed by mass spectrometry. Note, that lanes containing non-cross-linked, recombinant proteins exhibit degradation products of these proteins.
FIGURE 4.
RioK1 and pICln compete for binding to PRMT5. A, GST-tagged RioK1 (lane 2) and pICln (lane 3) or GST alone (lane 4), covalently cross-linked to GSH-Sepharose were incubated with HeLa extract in a pulldown assay. After extensive washing, the precipitated proteins were eluted from the matrix by sample buffer, resolved by SDS-PAGE, and visualized via Coomassie staining. Intervening lanes have been spliced out. Lane 1 shows protein standard (M). B, in a competition assay, the PRMT5 complex was immunoprecipitated with anti-WD45/MEP50 antibodies from HeLa cell extract. The purified complex was incubated with increasing amounts (0.1 to 10 μg) of GST-tagged RioK1 (aa 1–242, left panel) or pICln (right panel). The complex composition was afterward analyzed by Western blotting using the indicated antibodies. C, HeLa cell extract was incubated with GST (20 μg, upper panel), GST-RioK1 (20 μg, middle panel), or GST-pICln (<20 μg, lower panel) and was afterward fractionated by size exclusion chromatography. The collected fractions were analyzed by Western blotting with the indicated antibodies. D, GST-tagged pICln (lanes 2–4) or GST (lanes 5–7) immobilized on glutathione-Sepharose were incubated with in vitro translated, [35S]methionine-labeled, full-length PRMT5 or truncations thereof in an interaction assay. After extensive washing, the precipitated proteins were eluted from the matrix in loading buffer, resolved by SDS-PAGE, Coomassie stained, and visualized by autoradiography. For comparison, 10% of in vitro translated proteins were loaded (lanes 8–10). E, schematic overview of the interacting domains of PRMT5, RioK1, and pICln. Interactions are indicated by arrows. Interacting domains are colored black. fl, full length; nt, N-terminal; ct, C-terminal; TCL, total cell lysate.
FIGURE 5.
RioK1 interacts with the PRMT5 methylation substrate nucleolin. A, the PRMT5 complex was immunoprecipitated (IP) from HeLa lysate with the indicated antibodies and incubated with recombinant SmD1/D2 in a methylation assay with [3H]_S_-adenosyl methionine at 37 °C for 2 h. The reaction was stopped by the addition of sample buffer, and the eluted proteins were resolved by SDS-PAGE and Western blotting. HeLa total cell lysate (TCL) was used as control. Amido Black staining of the PVDF membrane was used to visualize equal loading of SmD1/D2. The identity of the precipitated proteins was ensured via immunoblotting. The methylated species of SmD1/D2 were visualized via autoradiography. Normal rabbit serum (NRS) was used as control for immunoprecipitation. B, GST-tagged members of the PRMT5 complex were incubated with in vitro translated, [35S]methionine-labeled nucleolin in an interaction assay (lanes 2–7). The precipitated proteins were visualized by autoradiography and Coomassie staining. For comparison, 10% of in vitro translated nucleolin was loaded (lane 8). Note that bands visible in the Coomassie staining below the expected molecular mass of the recombinant, GST-tagged proteins represent degradation products of these proteins. Lane 1 shows protein standard (M). C, nucleolin was specifically immunoprecipitated from HEK293T extract, and the precipitates were analyzed for the presence of PRMT5 complex components by immunoblotting with the respective specific antibodies (lane 2). Normal mouse serum (NMS) was used for control immunoprecipitation (lane 3). Lane 1 shows total extract of HEK293T. D, in an in vitro methylation assay of GST-SmD1 (lane 2), SmD1/D2 heterodimer (lane 3), the RG-boxes of NFAR (aa 476–2102, lane 4), hnRNP U (aa 677–824, lane 5), nucleolin (aa 648–710, lane 6), or GST (lane 7) were incubated with [3H]_S_-adenosyl methionine. The reaction was stopped by the addition of sample buffer and resolved by SDS-PAGE. The methylated proteins were visualized by autoradiography. Note that bands visible in the Coomassie staining below the expected molecular weight of the recombinant, GST-tagged proteins represent degradation products of these proteins. E, HEK293T cells were treated with siRNAs against RioK1 and harvested at the indicated time points and analyzed by immunoblotting for indicated proteins. F, cell extracts obtained in E were used for antinucleolin immunoprecipitations and analyzed for the presence of symmetrically methylated nucleolin by immunoblotting with SYM10 antibody.
FIGURE 6.
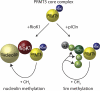
Model of PRMT5 substrate recognition through RioK1 and pICln. Schematic view of the mutually exclusive interaction of PRMT5 with RioK1 or pICln, which results in either the recruitment of nucleolin by RioK1 or Sm proteins by pICln for subsequent methylation by the PRMT5 complex.
Similar articles
- Molecular basis for substrate recruitment to the PRMT5 methylosome.
Mulvaney KM, Blomquist C, Acharya N, Li R, Ranaghan MJ, O'Keefe M, Rodriguez DJ, Young MJ, Kesar D, Pal D, Stokes M, Nelson AJ, Jain SS, Yang A, Mullin-Bernstein Z, Columbus J, Bozal FK, Skepner A, Raymond D, LaRussa S, McKinney DC, Freyzon Y, Baidi Y, Porter D, Aguirre AJ, Ianari A, McMillan B, Sellers WR. Mulvaney KM, et al. Mol Cell. 2021 Sep 2;81(17):3481-3495.e7. doi: 10.1016/j.molcel.2021.07.019. Epub 2021 Aug 5. Mol Cell. 2021. PMID: 34358446 Free PMC article. - Biochemical Investigation of the Interaction of pICln, RioK1 and COPR5 with the PRMT5-MEP50 Complex.
Krzyzanowski A, Gasper R, Adihou H, Hart P', Waldmann H. Krzyzanowski A, et al. Chembiochem. 2021 Jun 2;22(11):1908-1914. doi: 10.1002/cbic.202100079. Epub 2021 Mar 31. Chembiochem. 2021. PMID: 33624332 Free PMC article. - NF90/NFAR (nuclear factors associated with dsRNA) - a new methylation substrate of the PRMT5-WD45-RioK1 complex.
Cox J, Esser LM, Jüdt M, Schmitz K, Reiffert K, Grimmler M, Stork B, Wesselborg S, Peter C. Cox J, et al. Biol Chem. 2022 Aug 31;403(10):907-915. doi: 10.1515/hsz-2022-0136. Print 2022 Sep 27. Biol Chem. 2022. PMID: 36040368 - The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond.
Stopa N, Krebs JE, Shechter D. Stopa N, et al. Cell Mol Life Sci. 2015 Jun;72(11):2041-59. doi: 10.1007/s00018-015-1847-9. Epub 2015 Feb 7. Cell Mol Life Sci. 2015. PMID: 25662273 Free PMC article. Review. - The Structure and Function of the PRMT5:MEP50 Complex.
Antonysamy S. Antonysamy S. Subcell Biochem. 2017;83:185-194. doi: 10.1007/978-3-319-46503-6_7. Subcell Biochem. 2017. PMID: 28271477 Review.
Cited by
- Molecular basis for substrate recruitment to the PRMT5 methylosome.
Mulvaney KM, Blomquist C, Acharya N, Li R, Ranaghan MJ, O'Keefe M, Rodriguez DJ, Young MJ, Kesar D, Pal D, Stokes M, Nelson AJ, Jain SS, Yang A, Mullin-Bernstein Z, Columbus J, Bozal FK, Skepner A, Raymond D, LaRussa S, McKinney DC, Freyzon Y, Baidi Y, Porter D, Aguirre AJ, Ianari A, McMillan B, Sellers WR. Mulvaney KM, et al. Mol Cell. 2021 Sep 2;81(17):3481-3495.e7. doi: 10.1016/j.molcel.2021.07.019. Epub 2021 Aug 5. Mol Cell. 2021. PMID: 34358446 Free PMC article. - Structural insights into protein arginine symmetric dimethylation by PRMT5.
Sun L, Wang M, Lv Z, Yang N, Liu Y, Bao S, Gong W, Xu RM. Sun L, et al. Proc Natl Acad Sci U S A. 2011 Dec 20;108(51):20538-43. doi: 10.1073/pnas.1106946108. Epub 2011 Dec 5. Proc Natl Acad Sci U S A. 2011. PMID: 22143770 Free PMC article. - Aberrant expression of CITED2 promotes prostate cancer metastasis by activating the nucleolin-AKT pathway.
Shin SH, Lee GY, Lee M, Kang J, Shin HW, Chun YS, Park JW. Shin SH, et al. Nat Commun. 2018 Oct 5;9(1):4113. doi: 10.1038/s41467-018-06606-2. Nat Commun. 2018. PMID: 30291252 Free PMC article. - Regulation of a PRMT5/NF-κB Axis by Phosphorylation of PRMT5 at Serine 15 in Colorectal Cancer.
Hartley AV, Wang B, Jiang G, Wei H, Sun M, Prabhu L, Martin M, Safa A, Sun S, Liu Y, Lu T. Hartley AV, et al. Int J Mol Sci. 2020 May 23;21(10):3684. doi: 10.3390/ijms21103684. Int J Mol Sci. 2020. PMID: 32456215 Free PMC article. - Protein Arginine Methyltransferase 5 Functions via Interacting Proteins.
Liang Z, Wen C, Jiang H, Ma S, Liu X. Liang Z, et al. Front Cell Dev Biol. 2021 Aug 27;9:725301. doi: 10.3389/fcell.2021.725301. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34513846 Free PMC article.
References
- Gary J. D., Clarke S. (1998) Prog. Nucleic Acid Res. Mol. Biol. 61, 65–131 - PubMed
- Bedford M. T., Richard S. (2005) Mol. Cell 18, 263–272 - PubMed
- Branscombe T. L., Frankel A., Lee J. H., Cook J. R., Yang Z., Pestka S., Clarke S. (2001) J. Biol. Chem. 276, 32971–32976 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases