Activation of STIM1-Orai1 involves an intramolecular switching mechanism - PubMed (original) (raw)
Activation of STIM1-Orai1 involves an intramolecular switching mechanism
Marek K Korzeniowski et al. Sci Signal. 2010.
Abstract
Stromal interaction molecule 1 (STIM1) stimulates calcium ion (Ca(2+)) entry through plasma membrane Orai1 channels in response to decreased Ca(2+) concentrations in the endoplasmic reticulum lumen. We identified an acidic motif within the STIM1 coiled-coil region that keeps its Ca(2+) activation domain [Ca(2+) release-activated Ca(2+) (CRAC) activation domain/STIM1-Orai activating region (CAD/SOAR)]-a cytoplasmic region required for its activation of Orai1-inactive. The sequence of the STIM1 acidic motif shows substantial similarity to that of the carboxyl-terminal coiled-coil segment of Orai1, which is the postulated site of interaction with STIM1. Mutations within this acidic region rendered STIM1 constitutively active, whereas mutations within a short basic segment of CAD/SOAR prevented Orai1 activation. We propose that the CAD/SOAR domain is released from an intramolecular clamp during STIM1 activation, allowing the basic segment to activate Orai1 channels. This evolutionarily conserved mechanism of STIM1 activation resembles the regulation of protein kinases by intramolecular silencing through pseudosubstrate binding.
Figures
Figure 1
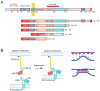
Clustering of the cytosolic domain of STIM1 activates Orai1 channels. (A) Schematics of STIM1 structure and the constructs used in the present study. The numbering corresponds to the human STIM1 protein. EF, Ca2+ binding EF-hand motif; SAM, sterile alpha motif; CAD/SOAR minimal Orai1 activation domain; D, acidic region; S/P, proline-, serine/threonine-rich segment, K, polybasic domain (after(39). The blue and yellow asterisks indicate the positions of the basic and acidic regions, respectively, identified in the present study. (B) Schematics of clustering by rapamycin-induced heterodimerization of the FKBP12 fused cytosolic STIM1 (FK-STIM1-ct) and the FRB targeted to the ER surface. The blue oval represents the C-terminal polybasic domain of STIM1 that is important for the PM localization of the protein (C) Localization of the indicated proteins expressed in COS-7 cells before (top row) and 5 min after (bottom row) rapamycin addition. Confocal images were taken in live cells 1 day after transfection. The area in the white box is shown enlarged. Note the significant membrane localization of the cytoplasmic STIM1 segment and its co-clustering with the Orai1 at the ER-PM contact zones after addition of rapamycin (100 nM). (D) Rapamycin-induced clustering increases FRET between YFP- and mRFP-tagged recruitable STIM1-ct indicating that clustering forces them to get within FRET distance. (E) Cytosolic Ca2+ increases evoked by rapamycin-induced clustering of FK-STIM1-ct. COS-7 cells were transfected with the ER-targeted CFP-FRB, the mRFP-FKBP12-STIM1-ct and untagged Orai1. Cytosolic Ca2+ changes were followed by Fura2. Blue represent cells with no visible transfections. Means ± S.E.M. are shown (n=51 and 58 cells for red and blue, respectively, form two separate experiments).
Figure 1
Clustering of the cytosolic domain of STIM1 activates Orai1 channels. (A) Schematics of STIM1 structure and the constructs used in the present study. The numbering corresponds to the human STIM1 protein. EF, Ca2+ binding EF-hand motif; SAM, sterile alpha motif; CAD/SOAR minimal Orai1 activation domain; D, acidic region; S/P, proline-, serine/threonine-rich segment, K, polybasic domain (after(39). The blue and yellow asterisks indicate the positions of the basic and acidic regions, respectively, identified in the present study. (B) Schematics of clustering by rapamycin-induced heterodimerization of the FKBP12 fused cytosolic STIM1 (FK-STIM1-ct) and the FRB targeted to the ER surface. The blue oval represents the C-terminal polybasic domain of STIM1 that is important for the PM localization of the protein (C) Localization of the indicated proteins expressed in COS-7 cells before (top row) and 5 min after (bottom row) rapamycin addition. Confocal images were taken in live cells 1 day after transfection. The area in the white box is shown enlarged. Note the significant membrane localization of the cytoplasmic STIM1 segment and its co-clustering with the Orai1 at the ER-PM contact zones after addition of rapamycin (100 nM). (D) Rapamycin-induced clustering increases FRET between YFP- and mRFP-tagged recruitable STIM1-ct indicating that clustering forces them to get within FRET distance. (E) Cytosolic Ca2+ increases evoked by rapamycin-induced clustering of FK-STIM1-ct. COS-7 cells were transfected with the ER-targeted CFP-FRB, the mRFP-FKBP12-STIM1-ct and untagged Orai1. Cytosolic Ca2+ changes were followed by Fura2. Blue represent cells with no visible transfections. Means ± S.E.M. are shown (n=51 and 58 cells for red and blue, respectively, form two separate experiments).
Figure 2
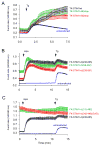
Cytoplasmic Ca2+ responses evoked by various STIM1 constructs truncated from the C-terminus. (A) C-terminal truncations on the full-STIM1 (with intact N-terminus) protein up to residue 463 have only small effects on the thapsigargin (Tg, 200 nM) –induced Ca2+ elevations. Cells were co-transfected with untagged Orai1 and the indicated mRFP-tagged (in the luminal side) STIM1 constructs driven by the thimidine kinase promoter (TK-STIM1). Means ± S.E.M. are shown (n = 89, 37, 32 and 198 cells for black, green, red and blue traces, respectively obtained in 2–5 separate experiments). (B) The same truncations have significant impact on the abilities of the cytoplasmic STIM1 (STIM1-ct) pieces to activate Orai1 channels after rapamycin-induced clustering. Here the indicated STIM1-ct fragments were fused with the mRFP-FKBP12 module (FK-STIM-ct) and co-expressed with untagged Orai1 and the ER-targeted CFP-FRB. Means ± S.E.M. are shown (n = 356, 254, 259 and 639 cells for black, green, red and blue traces, respectively obtained in 9–10 separate experiments). (C) Truncation of the cytoplasmic STIM1 fragments from the N-terminal direction makes them constitutively active not requiring clustering. Cells were transfected with the indicated constructs as described in panel B. Means ± S.E.M. are shown (n = 208, 82, 124 and 305 cells for black, green, red and blue traces, respectively obtained in 4–6 separate experiments).
Figure 3
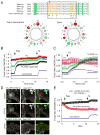
An acidic segment within the coiled-coil domain of STIM1 is homologous with the Orai1 C-terminal helix and is essential to keep STIM1 in an inactive state. (A) Sequence alignment between STIM1 orthologues and the Orai1 C-terminus identifies a homologous region within the coiled-coil domain. Helical wheel diagram shows the high degree of positional conservation between the Orai1 helix and the proximal part of the acidic STIM1 segment. A Leu to Ser substitution of the residue shown by the red arrow in Orai1 has been shown to prevent its activation by STIM1 (22). Mutations within this segment of STIM1 are labeled by an asterisk (AGAA, or 3EA). Changing only the EL residues in STIM1 (to AG) had negligible effects (not shown). (B) Mutations within this segment (3EA) make the cytoplasmic STIM1 construct partially active without clustering (red). However, mutation of the four E residues in the distal part of the acidic motif (4EA) renders the cytoplasmic STIM1 piece fully constitutively fully active. Means ± S.E.M. are shown (n = 220, 345, 143 and 802 cells for black, green, red and blue traces, respectively obtained in 6–38 separate dishes from 3–9 independent experiment). (C) The same mutations within the full-length STIM1 protein show similar partial or full constitutive activity. For comparison, the effects of the known constitutively active D76A mutant (4) STIM1 is shown. For these experiments the thymidine kinase (TK) driven mRFP-STIM1 constructs were expressed together with untagged Orai1. Means ± S.E.M. are shown (n = 143, 109, 80, 111 and 785 cells for black, green, pink, red and blue traces, respectively, obtained in 3–36 dishes from 3–8 separate experiments). (D) Unlike wild-type STIM1 (upper two rows), the 4EA mutant mRFP-STIM1 forms constitutive clusters together with Orai1-YFP without Ca2+ release from the ER (lower row). This analysis was performed at room temperature. (E) Plasma membrane recruitment of the STIM1 coiled-coil domain containing the inhibitory acidic segment inhibits Orai1-mediated Ca2+ influx activated by the constitutively active STIM1 fragment (315–462). For this experiments COS-7 cells were transfected with the mRFP-fused STIM1(315–462) (non-recruitable!), untagged Orai1 and the ER-recruiter FRB together with either the mRFP-FKBP12-fused STIM1-ct(238–343) [FK-STIM1-ct (238–443), red] or mRFP-FKBPonly (FK-only, black). Rapamycin addition decreased the high basal Ca2+ in a significant fraction of cells (~16 %) in the STIM1-ct(238–343) expressing group while had no effect in any cells in the FKBPonly group. Means ± S.E.M. are shown (n = 119, 80 and 247 cells for black, red and blue traces, respectively, obtained in 5–10 separate experiments).
Figure 4
A polybasic segment within the CAD/SOAR domain is responsible for Orai1 activation. (A) sequence alignment between various STIM1 orthologues and STIM2 within the CAD/SOAR domain (gg, Gallus gallus; xl, Xenopus laevis; dr, danio rero; dm, Drosophila melanogaster. The mutated Lys residues are labeled with an asterisk. (B) The 4K mutation renders full-length STIM1 proteins inactive. COS-7 cells were transfected with the thimidine-kinase (TK) driven STIM1 (black) or its 4K mutant (red) together with untagged Orai1 and their cytoplasmic Ca2+ monitored by Fura2. Thapsigargin (Tg, 200 nM) was added to empty the Ca2+ stores. Means ± S.E.M. are shown (n = 135, 132 and 160 cells for black, red and blue traces, respectively, obtained in 4–5 separate experiments). (C) The same 4K mutation makes the STIM1-ct fragment inactive not responding to clustering with Ca2+ increase (red). Means ± S.E.M. are shown (n = 170, 189 and 239 cells for black, red and blue traces, respectively obtained in 6–7 separate experiments). (D) The 4K(AGAG) mutation inactivates the otherwise constitutively active STIM1-ct(315–462) segment. Means ± S.E.M. are shown (n = 188, 163 and 244 cells for black, red and blue traces, respectively, obtained in 5–7 separate experiments). (E) The constitutively active STIM1(315–462) piece causes clustering and co-localization with Orai1 in COS-7 cells. However, the 4K mutation prevents clustering of Orai1 and the STIM1 construct is largely remains in the cytosol. Confocal images of live cells are shown one day after transfection.
Similar articles
- A basic sequence in STIM1 promotes Ca2+ influx by interacting with the C-terminal acidic coiled coil of Orai1.
Calloway N, Holowka D, Baird B. Calloway N, et al. Biochemistry. 2010 Feb 16;49(6):1067-71. doi: 10.1021/bi901936q. Biochemistry. 2010. PMID: 20073506 Free PMC article. - Defects in the STIM1 SOARα2 domain affect multiple steps in the CRAC channel activation cascade.
Höglinger C, Grabmayr H, Maltan L, Horvath F, Krobath H, Muik M, Tiffner A, Renger T, Romanin C, Fahrner M, Derler I. Höglinger C, et al. Cell Mol Life Sci. 2021 Oct;78(19-20):6645-6667. doi: 10.1007/s00018-021-03933-4. Epub 2021 Sep 8. Cell Mol Life Sci. 2021. PMID: 34498097 Free PMC article. - SOAR and the polybasic STIM1 domains gate and regulate Orai channels.
Yuan JP, Zeng W, Dorwart MR, Choi YJ, Worley PF, Muallem S. Yuan JP, et al. Nat Cell Biol. 2009 Mar;11(3):337-43. doi: 10.1038/ncb1842. Epub 2009 Feb 1. Nat Cell Biol. 2009. PMID: 19182790 Free PMC article. - Modulation of Orai1 and STIM1 by Cellular Factors.
Woo JS, Srikanth S, Gwack Y. Woo JS, et al. In: Kozak JA, Putney JW Jr, editors. Calcium Entry Channels in Non-Excitable Cells. Boca Raton (FL): CRC Press/Taylor & Francis; 2018. Chapter 4. In: Kozak JA, Putney JW Jr, editors. Calcium Entry Channels in Non-Excitable Cells. Boca Raton (FL): CRC Press/Taylor & Francis; 2018. Chapter 4. PMID: 30299655 Free Books & Documents. Review. - Store-Independent Orai Channels Regulated by STIM.
Zhang X, Gueguinou M, Trebak M. Zhang X, et al. In: Kozak JA, Putney JW Jr, editors. Calcium Entry Channels in Non-Excitable Cells. Boca Raton (FL): CRC Press/Taylor & Francis; 2018. Chapter 11. In: Kozak JA, Putney JW Jr, editors. Calcium Entry Channels in Non-Excitable Cells. Boca Raton (FL): CRC Press/Taylor & Francis; 2018. Chapter 11. PMID: 30299650 Free Books & Documents. Review.
Cited by
- Synthetic Biology Meets Ca2+ Release-Activated Ca2+ Channel-Dependent Immunomodulation.
Bacsa B, Hopl V, Derler I. Bacsa B, et al. Cells. 2024 Mar 7;13(6):468. doi: 10.3390/cells13060468. Cells. 2024. PMID: 38534312 Free PMC article. Review. - STIM1 translocation to the nucleus protects cells from DNA damage.
Sanchez-Lopez I, Orantos-Aguilera Y, Pozo-Guisado E, Alvarez-Barrientos A, Lilla S, Zanivan S, Lachaud C, Martin-Romero FJ. Sanchez-Lopez I, et al. Nucleic Acids Res. 2024 Mar 21;52(5):2389-2415. doi: 10.1093/nar/gkae001. Nucleic Acids Res. 2024. PMID: 38224453 Free PMC article. - Molecular Mechanism Analysis of STIM1 Thermal Sensation.
Liu X, Zheng T, Jiang Y, Wang L, Zhang Y, Liang Q, Chen Y. Liu X, et al. Cells. 2023 Nov 12;12(22):2613. doi: 10.3390/cells12222613. Cells. 2023. PMID: 37998348 Free PMC article. - An apical Phe-His pair defines the Orai1-coupling site and its occlusion within STIM1.
Zhou Y, Jennette MR, Ma G, Kazzaz SA, Baraniak JH, Nwokonko RM, Groff ML, Velasquez-Reynel M, Huang Y, Wang Y, Gill DL. Zhou Y, et al. Nat Commun. 2023 Oct 30;14(1):6921. doi: 10.1038/s41467-023-42254-x. Nat Commun. 2023. PMID: 37903816 Free PMC article. - S417 in the CC3 region of STIM1 is critical for STIM1-Orai1 binding and CRAC channel activation.
Yu T, Li X, Luo Q, Liu H, Jin J, Li S, He J. Yu T, et al. Life Sci Alliance. 2023 Jan 23;6(4):e202201623. doi: 10.26508/lsa.202201623. Print 2023 Apr. Life Sci Alliance. 2023. PMID: 36690443 Free PMC article.
References
- Putney JW., Jr A model for receptor-regulated calcium entry. Cell Calcium. 1986;7:1–12. - PubMed
- Hoth M, Penner R. Depletion of intracellular calcium stores activates a calcium current in mast cells. Nature. 1992;355:353–356. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous