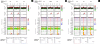An initial comparative map of copy number variations in the goat (Capra hircus) genome - PubMed (original) (raw)
Comparative Study
An initial comparative map of copy number variations in the goat (Capra hircus) genome
Luca Fontanesi et al. BMC Genomics. 2010.
Abstract
Background: The goat (Capra hircus) represents one of the most important farm animal species. It is reared in all continents with an estimated world population of about 800 million of animals. Despite its importance, studies on the goat genome are still in their infancy compared to those in other farm animal species. Comparative mapping between cattle and goat showed only a few rearrangements in agreement with the similarity of chromosome banding. We carried out a cross species cattle-goat array comparative genome hybridization (aCGH) experiment in order to identify copy number variations (CNVs) in the goat genome analysing animals of different breeds (Saanen, Camosciata delle Alpi, Girgentana, and Murciano-Granadina) using a tiling oligonucleotide array with ~385,000 probes designed on the bovine genome.
Results: We identified a total of 161 CNVs (an average of 17.9 CNVs per goat), with the largest number in the Saanen breed and the lowest in the Camosciata delle Alpi goat. By aggregating overlapping CNVs identified in different animals we determined CNV regions (CNVRs): on the whole, we identified 127 CNVRs covering about 11.47 Mb of the virtual goat genome referred to the bovine genome (0.435% of the latter genome). These 127 CNVRs included 86 loss and 41 gain and ranged from about 24 kb to about 1.07 Mb with a mean and median equal to 90,292 bp and 49,530 bp, respectively. To evaluate whether the identified goat CNVRs overlap with those reported in the cattle genome, we compared our results with those obtained in four independent cattle experiments. Overlapping between goat and cattle CNVRs was highly significant (P < 0.0001) suggesting that several chromosome regions might contain recurrent interspecies CNVRs. Genes with environmental functions were over-represented in goat CNVRs as reported in other mammals.
Conclusions: We describe a first map of goat CNVRs. This provides information on a comparative basis with the cattle genome by identifying putative recurrent interspecies CNVs between these two ruminant species. Several goat CNVs affect genes with important biological functions. Further studies are needed to evaluate the functional relevance of these CNVs and their effects on behavior, production, and disease resistance traits in goats.
Figures
Figure 1
Goat of different breeds used for CNVs discovery. A = Saanen; B = Camosciata delle Alpi; C = Girgentana; D = Murciano-Granadina.
Figure 2
Comparative map of CNVRs identified in goats reported on the bovine chromosomes.
Figure 3
aCGH and SQF-PCR results for CNVRs nos. 25 (A), 61-62 (B), and 90 (C). For each CNVR, results were reported for three goats indicated at the top in the correspondence of the related charts and images (C1 = Camosciata delle Alpi, animal no. 1; G1 = Girgentana, animal no. 1; G2 = Girgentana, animal no. 2; MG1 = Murciano-Granadina, animal no. 1; S1 = Saanen, animal no. 1; S3 = Saanen, animal no. 3). For the aCGH experiment, images have been reported for i) log2 ratio plot of original data, ii) log2 ratio plot of summary data (pointwise averaging of all computed profiles), iii) heatmap of log2 ratios for original, smoothed/segmented, and summary data, and iv) maps of gains/losses for smoothed/segmented and summary data (gain is indicated in orange, loss is indicated in green). Red arrows indicate regions of copy gain. Smoothed/segmented data were obtained with several algorithms (Lowess, Wavelet, Quantreg, ruavg, CBS, CGHseg, BioHMM, cghFLasso, GLAD, and FASeg) averaged in the summary data [77].. Semiquantitative fluorescent multiplex PCR (SQF-PCR) electropherograms for fragments of three CNVRs are superimposed on that of the reference Camosciata delle Alpi goat after normalization against the control MC1R amplicon. Results obtained using DGAT1 for normalization are overlapping with those obtained with MC1R and for this reason are not reported.
Similar articles
- A first comparative map of copy number variations in the sheep genome.
Fontanesi L, Beretti F, Martelli PL, Colombo M, Dall'olio S, Occidente M, Portolano B, Casadio R, Matassino D, Russo V. Fontanesi L, et al. Genomics. 2011 Mar;97(3):158-65. doi: 10.1016/j.ygeno.2010.11.005. Epub 2010 Nov 24. Genomics. 2011. PMID: 21111040 - Copy number variation and missense mutations of the agouti signaling protein (ASIP) gene in goat breeds with different coat colors.
Fontanesi L, Beretti F, Riggio V, Gómez González E, Dall'Olio S, Davoli R, Russo V, Portolano B. Fontanesi L, et al. Cytogenet Genome Res. 2009;126(4):333-47. doi: 10.1159/000268089. Epub 2009 Dec 16. Cytogenet Genome Res. 2009. PMID: 20016133 - Identification and population genetic analyses of copy number variations in six domestic goat breeds and Bezoar ibexes using next-generation sequencing.
Guo J, Zhong J, Liu GE, Yang L, Li L, Chen G, Song T, Zhang H. Guo J, et al. BMC Genomics. 2020 Nov 27;21(1):840. doi: 10.1186/s12864-020-07267-6. BMC Genomics. 2020. PMID: 33246410 Free PMC article. - Genome wide copy number variations using Porcine 60K SNP Beadchip in Landlly pigs.
Panda S, Kumar A, Gaur GK, Ahmad SF, Chauhan A, Mehrotra A, Dutt T. Panda S, et al. Anim Biotechnol. 2023 Nov;34(6):1891-1899. doi: 10.1080/10495398.2022.2056047. Epub 2022 Apr 4. Anim Biotechnol. 2023. PMID: 35369845 Review. - Molecular cytogenetics and comparative mapping in goats (Capra hircus, 2n = 60).
Schibler L, Di Meo GP, Cribiu EP, Iannuzzi L. Schibler L, et al. Cytogenet Genome Res. 2009;126(1-2):77-85. doi: 10.1159/000245908. Epub 2009 Dec 9. Cytogenet Genome Res. 2009. PMID: 20016158 Review.
Cited by
- Adaptive potential of genomic structural variation in human and mammalian evolution.
Radke DW, Lee C. Radke DW, et al. Brief Funct Genomics. 2015 Sep;14(5):358-68. doi: 10.1093/bfgp/elv019. Epub 2015 May 23. Brief Funct Genomics. 2015. PMID: 26003631 Free PMC article. Review. - Genome-wide patterns of copy number variation in the Chinese yak genome.
Zhang X, Wang K, Wang L, Yang Y, Ni Z, Xie X, Shao X, Han J, Wan D, Qiu Q. Zhang X, et al. BMC Genomics. 2016 May 20;17:379. doi: 10.1186/s12864-016-2702-6. BMC Genomics. 2016. PMID: 27206476 Free PMC article. - Optimization of Saanen sperm genes amplification: evaluation of standardized protocols in genetically uncharacterized rural goats reared under a subtropical environment.
Barbour EK, Saade MF, Sleiman FT, Hamadeh SK, Mouneimne Y, Kassaifi Z, Kayali G, Harakeh S, Jaber LS, Shaib HA. Barbour EK, et al. Trop Anim Health Prod. 2012 Oct;44(7):1513-9. doi: 10.1007/s11250-012-0096-2. Epub 2012 Feb 18. Trop Anim Health Prod. 2012. PMID: 22350811 - Next Generation Semiconductor Based Sequencing of the Donkey (Equus asinus) Genome Provided Comparative Sequence Data against the Horse Genome and a Few Millions of Single Nucleotide Polymorphisms.
Bertolini F, Scimone C, Geraci C, Schiavo G, Utzeri VJ, Chiofalo V, Fontanesi L. Bertolini F, et al. PLoS One. 2015 Jul 7;10(7):e0131925. doi: 10.1371/journal.pone.0131925. eCollection 2015. PLoS One. 2015. PMID: 26151450 Free PMC article. - The application of genome-wide SNP genotyping methods in studies on livestock genomes.
Gurgul A, Semik E, Pawlina K, Szmatoła T, Jasielczuk I, Bugno-Poniewierska M. Gurgul A, et al. J Appl Genet. 2014 May;55(2):197-208. doi: 10.1007/s13353-014-0202-4. Epub 2014 Feb 25. J Appl Genet. 2014. PMID: 24566962 Review.
References
- FAO. The State of the World's Animal Genetic Resources for Food and Agriculture, Rome. 2007.
- Schibler L, Vaiman D, Oustry A, Giraud-Delville C, Cribiu EP. Comparative gene mapping: a fine-scale survey of chromosome rearrangements between ruminants and humans. Genome Res. 1998;8:901–915. - PubMed
- Gatesy J, Yelon D, DeSalle R, Vrba ES. Phylogeny of the Bovidae (Artiodactyla, Mammalia), based on mitochondrial ribosomal DNA sequences. Mol Biol Evol. 1992;9:433–446. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous