The SNO-proteome: causation and classifications - PubMed (original) (raw)
Review
The SNO-proteome: causation and classifications
Divya Seth et al. Curr Opin Chem Biol. 2011 Feb.
Abstract
Cell signaling is a complex and highly regulated process. Post-translational modifications of proteins serve to sense and transduce cellular signals in a precisely coordinated manner. It is increasingly recognized that protein S-nitrosylation, the addition of a nitric oxide group to cysteine thiols, serves an important role in a wide range of signaling pathways. In spite of the large number of SNO-proteins now identified (∼1000), the observed specificity of S-nitrosylation in terms of target proteins and specific cysteines within modified proteins is incompletely understood. Here we review the progress made in S-nitrosylation detection methods that have facilitated the study of the SNO-proteome under physiological and pathophysiological conditions, and some factors important in determining the SNO-proteome. Classification schemes for emergent denitrosylases and prospective 'protein S-nitrosylases' are provided.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures
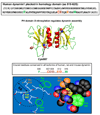
Figure 1. _S_-nitrosylation of a crucial cysteine in the pleckstrin homology domain regulates dynamin multimerization
The _S_-nitrosylated Cys607 lies in a hydrophobic pocket (so-called “hydrophobic core” of the pleckstrin homology domain), which contains aromatic side-chains and an acid-base motif within a 6Å radius (“acid-base/hydrophobic” motif). The residues that form the proposed S-nitrosylation motif are conserved in mammalian dynamins and are highlighted in the pleckstrin homology domain sequence.
Similar articles
- Protein S-Nitrosylation: Determinants of Specificity and Enzymatic Regulation of S-Nitrosothiol-Based Signaling.
Stomberski CT, Hess DT, Stamler JS. Stomberski CT, et al. Antioxid Redox Signal. 2019 Apr 1;30(10):1331-1351. doi: 10.1089/ars.2017.7403. Epub 2018 Jan 10. Antioxid Redox Signal. 2019. PMID: 29130312 Free PMC article. Review. - Molecular recognition of _S_-nitrosothiol substrate by its cognate protein denitrosylase.
Stomberski CT, Zhou HL, Wang L, van den Akker F, Stamler JS. Stomberski CT, et al. J Biol Chem. 2019 Feb 1;294(5):1568-1578. doi: 10.1074/jbc.RA118.004947. Epub 2018 Dec 11. J Biol Chem. 2019. PMID: 30538128 Free PMC article. - Proteome-wide detection of S-nitrosylation targets and motifs using bioorthogonal cleavable-linker-based enrichment and switch technique.
Mnatsakanyan R, Markoutsa S, Walbrunn K, Roos A, Verhelst SHL, Zahedi RP. Mnatsakanyan R, et al. Nat Commun. 2019 May 16;10(1):2195. doi: 10.1038/s41467-019-10182-4. Nat Commun. 2019. PMID: 31097712 Free PMC article. - Proteome-wide modulation of S-nitrosylation in Trypanosoma cruzi trypomastigotes upon interaction with the host extracellular matrix.
Mule SN, Manchola NC, de Oliveira GS, Pereira M, Magalhães RDM, Teixeira AA, Colli W, Alves MJM, Palmisano G. Mule SN, et al. J Proteomics. 2021 Jan 16;231:104020. doi: 10.1016/j.jprot.2020.104020. Epub 2020 Oct 20. J Proteomics. 2021. PMID: 33096306 - Proteomic approaches to evaluate protein S-nitrosylation in disease.
López-Sánchez LM, López-Pedrera C, Rodríguez-Ariza A. López-Sánchez LM, et al. Mass Spectrom Rev. 2014 Jan-Feb;33(1):7-20. doi: 10.1002/mas.21373. Epub 2013 Jun 15. Mass Spectrom Rev. 2014. PMID: 23775552 Review.
Cited by
- Mechanisms of nitrosylation and denitrosylation of cytoplasmic glyceraldehyde-3-phosphate dehydrogenase from Arabidopsis thaliana.
Zaffagnini M, Morisse S, Bedhomme M, Marchand CH, Festa M, Rouhier N, Lemaire SD, Trost P. Zaffagnini M, et al. J Biol Chem. 2013 Aug 2;288(31):22777-89. doi: 10.1074/jbc.M113.475467. Epub 2013 Jun 7. J Biol Chem. 2013. PMID: 23749990 Free PMC article. - Repletion of S-nitrosohemoglobin improves organ function and physiological status in swine after brain death.
Yurcisin BM, Davison TE, Bibbs SM, Collins BH, Stamler JS, Reynolds JD. Yurcisin BM, et al. Ann Surg. 2013 May;257(5):971-7. doi: 10.1097/SLA.0b013e3182822c52. Ann Surg. 2013. PMID: 23360919 Free PMC article. - Specificity in S-nitrosylation: a short-range mechanism for NO signaling?
Martínez-Ruiz A, Araújo IM, Izquierdo-Álvarez A, Hernansanz-Agustín P, Lamas S, Serrador JM. Martínez-Ruiz A, et al. Antioxid Redox Signal. 2013 Oct 10;19(11):1220-35. doi: 10.1089/ars.2012.5066. Epub 2013 Jan 4. Antioxid Redox Signal. 2013. PMID: 23157283 Free PMC article. Review. - STAT3 regulation by S-nitrosylation: implication for inflammatory disease.
Kim J, Won JS, Singh AK, Sharma AK, Singh I. Kim J, et al. Antioxid Redox Signal. 2014 Jun 1;20(16):2514-27. doi: 10.1089/ars.2013.5223. Epub 2014 Feb 14. Antioxid Redox Signal. 2014. PMID: 24063605 Free PMC article. - The Role of Protein S-Nitrosylation in Protein Misfolding-Associated Diseases.
Ju YJ, Lee HW, Choi JW, Choi MS. Ju YJ, et al. Life (Basel). 2021 Jul 17;11(7):705. doi: 10.3390/life11070705. Life (Basel). 2021. PMID: 34357077 Free PMC article. Review.
References
- Hess DT, Matsumoto A, Kim S-O, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. - PubMed
- Ozawa K, Whalen EJ, Nelson CD, Mu Y, Hess DT, Lefkowitz RJ, Stamler JS. S-nitrosylation of beta-arrestin regulates beta-adrenergic receptor trafficking. Mol Cell. 2008;31:395–405. • The authors show that a crucial component of GPCR signaling, b-arrestin 2, interacts with and is S-nitrosylated at a single cysteine by endothelial NO synthase (eNOS).
- Xu L, Eu JP, Meissner G, Stamler JS. Activation of the cardiac calcium release channel (ryanodine receptor) by poly-S-nitrosylation. Science. 1998;279:234–237. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL091876/HL/NHLBI NIH HHS/United States
- R01 HL059130-13/HL/NHLBI NIH HHS/United States
- R01 HL095463-04/HL/NHLBI NIH HHS/United States
- R01 HL096673/HL/NHLBI NIH HHS/United States
- R01 HL095463/HL/NHLBI NIH HHS/United States
- R01 HL096673-02/HL/NHLBI NIH HHS/United States
- R01 HL059130/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources