Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity - PubMed (original) (raw)
Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity
Seung-Jae Lee et al. Curr Biol. 2010.
Abstract
A mild inhibition of mitochondrial respiration extends the life span of many organisms, including yeast, worms, flies, and mice, but the underlying mechanism is unknown. One environmental condition that reduces rates of respiration is hypoxia (low oxygen). Thus, it is possible that mechanisms that sense oxygen play a role in the longevity response to reduced respiration. The hypoxia-inducible factor HIF-1 is a highly conserved transcription factor that activates genes that promote survival during hypoxia. In this study, we show that inhibition of respiration in C. elegans can promote longevity by activating HIF-1. Through genome-wide screening, we found that RNA interference (RNAi) knockdown of many genes encoding respiratory-chain components induced hif-1-dependent transcription. Moreover, HIF-1 was required for the extended life spans of clk-1 and isp-1 mutants, which have reduced rates of respiration. Inhibiting respiration appears to activate HIF-1 by elevating the level of reactive oxygen species (ROS). We found that ROS are increased in respiration mutants and that mild increases in ROS can stimulate HIF-1 to activate gene expression and promote longevity. In this way, HIF-1 appears to link respiratory stress in the mitochondria to a nuclear transcriptional response that promotes longevity.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures
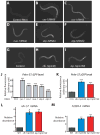
Fig. 1. Inhibiting respiration increases HIF-1 activity
(A) Animals expressing the HIF-1-regulated Pnhr-57::GFP transgene displayed low levels of GFP when grown on control bacteria carrying the empty RNAi vector. (B–F) In contrast, RNAi of respiratory-chain or ATP synthase genes cyc-1 (B), cco-1 (C), nuo-1 (D), atp-3 (E), or atp-5 (F) induced the expression of Pnhr-57::GFP. RNAi treatment of cyc-1 or cco-1 only during adulthood did not increase the level of Pnhr-57::GFP (Fig. S1D-H). (G–I) Mutations in clk-1 (H) and isp-1 (I), which reduce respiration, elevated Pnhr-57::GFP expression (G). (J) Quantification of fluorescence in A to F (n>30); and (K), in G to I (n>42). mRNA levels of nhr-57 (L) and F22B5.4 (M), another HIF-1-regulated gene, were significantly increased in clk-1(qm30) and isp-1(qm150) mutants [Please see Fig. S1A-C for qRT-PCR data of other hypoxia-responsive genes]. Data were obtained from 3 independent quantitative RT-PCR analyses and error bars represent s.e.m (*P<0.01, **P<0.001, ***P<0.0001, two-tailed Student’s _t_-test compared to wild type).
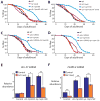
Fig. 2. The lifespan extension conferred by respiration mutants requires hif-1
(A, B) hif-1(ia4) loss-of-function mutations decreased the longevity of clk-1(qm30) (A) and isp-1(qm150) (B) mutants significantly (in three out of four trials and four out of four trials, respectively; See Supplemental Table S2). (C, D) The long lifespan of clk-1(qm30) (C) and isp-1(qm150) (D) mutants was significantly shortened by aha-1 [HIF1β] RNAi. Neither the hif-1(ia4) mutation nor aha-1 RNAi affected the lifespan of wild type (WT). (See Supplemental Table S2 for statistical analysis.) [We note that whereas Mehta et al. and we both found that hif-1 mutations do not affect the lifespans of wild type [19], Chen et al. and Zhang et al. reported that hif-1 mutants live longer than wild type [–38]. We carried out additional experiments to resolve this discrepancy, which are described in supplemental material (Fig. S2M, N)] (E, F) The increased mRNA levels of the HIF-1-dependent genes nhr-57 (E) and F22B5.4 (F) in clk-1(qm30) and isp-1(qm150) mutants were significantly decreased by hif-1(ia4) mutation. Error bars represent s.e.m (n=3, *P<0.05, **P<0.01, two-tailed Student’s _t_-test). See Fig. S2G, H for quantitative RT-PCR data of other _hif-1_-dependent hypoxia-inducible genes.
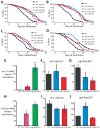
Fig. 3. Activation of HIF-1 by vhl-1 or egl-9 mutations does not further lengthen the lifespan of respiration mutants
(A, B) Mutations in vhl-1 increased the lifespan of wild type but did not further extend the lifespan of clk-1(qm30) (A) or isp-1(qm150) (B) mutants. (C, D) The egl-9(sa307) mutation did not further increase the lifespans of clk-1(qm30) (C) or isp-1(qm150) (D) mutants. (E) Consistent with previous reports [14, 39], mRNA levels of nhr-57 were significantly increased by vhl-1(ok161) and egl-9(sa307) mutations. (F, G) The increased mRNA levels of nhr-57 in vhl-1(ok161) (F) or egl-9(sa307) (G) mutants were not further augmented by clk-1(qm30) and isp-1(qm150) mutations (control, Ctrl.). (See Supplemental Table S3 for statistical analysis.) (H) Expression of F22B5.4 was highly induced in vhl-1(ok161) and egl-9(sa307) mutants as reported previously [14, 39]. (I, J) this induction was not significantly further increased by clk-1(qm30) and isp-1(qm150) mutations. Note that the increased F22B5.4 mRNA levels in egl-9(sa307) mutants by clk-1(qm30) mutation was marginally not significant (_P_=0.06). Error bars represent s.e.m (n=3, *P<0.01, **P<0.001, two-tailed Student’s _t_-test).
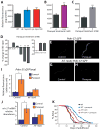
Fig. 4. Increased ROS causes HIF-1 to promote longevity in respiration-defective mutants
(A) ROS levels, measured using a 2′,7′-dichlorofluorescein diacetate (DCF-DA) fluorescence assay, were significantly increased in clk-1(qm30) and isp-1(qm150) mutants (n=5). See also Fig. S4A for data using mev-1(kn1) mutant animals, which were previously shown to have increased ROS levels [40]. (B–C) A low dose (0.25 mM) (B) and high dose (4 mM) (C) of paraquat significantly increased the level of DCF-DA fluorescence. 4 mM paraquat was introduced from L4 to day 3 of adulthood because worms arrest as larvae if treated with 4 mM paraquat from hatching (n=3). Error bars represent s.e.m (*P<0.05, **_P_<0.01, two-tailed Student’s _t_-test). (**D**) Low doses of paraquat (0.125 mM, 0.25 mM, 0.5 mM and 1 mM) lengthened lifespan, whereas higher concentrations (4, 16, and 64 mM) shortened lifespan. Paraquat was introduced during adulthood. The lifespan measurements for 0.125 mM and 0.5 mM paraquat treatment were performed separately and therefore shown with different controls. See also Table S4. (**E**, **F**) Compared to untreated control _Pnhr-57::GFP_ animals (**E**), animals treated with low levels of paraquat (0.25 mM) displayed increased GFP levels (**F**). Paraquat treatment further increased _Pnhr-57::GFP_ levels in _clk-1(qm30)_ and _isp-1(qm150)_ mutant animals (Fig. S4J, K), suggesting that the induction was not saturated by either of the mutations or by the paraquat treatment. (**G**, **H**) The induction of _Pnhr-57::GFP_ by paraquat treatment was significantly diminished in the _hif-1(ia4)_ mutant. (**I**) Quantification of fluorescence in **E** to **H** (n >15). (J) The increased nhr-57 mRNA abundance caused by paraquat treatment, assayed using quantitative RT-PCR, was significantly decreased by hif-1(ia4) mutation. Data analysis was done from 10 independent quantitative RT-PCR experiments. [In two of our data sets, the levels of nhr-57 mRNA in paraquat-treated wild-type animals were increased by a very-large 681 and 755 fold compared to those in control animals. We excluded these two datasets from our analysis by using rejection analysis of QP-test for outliers (confidence level: 0.99) [41]]. (K) Mutations in hif-1 significantly decreased the longevity caused by paraquat treatment. Some long-lived mutants require the daf-16/FOXO transcription factor gene for their longevity, but respiration mutants do not [1, 4, 10]. We found that 0.25 mM paraquat treatment increased the lifespan of daf-16(mu86) null mutants (Fig. S4L). See Supplemental Table S4 for statistical analysis. Error bars represent s.e.m (*P<0.05, **P<0.01, two-tailed Student’s _t_-test). Previously, Rea et al. observed an increased trend in protein carbonylation levels at doses of respiratory-chain RNAi that increased lifespan, and also at higher RNAi doses, which did not extend lifespan [17]. On this basis, they concluded that ROS did not play a role in this longevity. One way to reconcile their findings with ours is to suggest that a sharp reduction in respiration prevents lifespan extension in spite of elevated ROS because it compromises the animals’ health.
Similar articles
- Regulation of life span by mitochondrial respiration: the HIF-1 and ROS connection.
Hwang AB, Lee SJ. Hwang AB, et al. Aging (Albany NY). 2011 Mar;3(3):304-10. doi: 10.18632/aging.100292. Aging (Albany NY). 2011. PMID: 21389351 Free PMC article. Review. - Feedback regulation via AMPK and HIF-1 mediates ROS-dependent longevity in Caenorhabditis elegans.
Hwang AB, Ryu EA, Artan M, Chang HW, Kabir MH, Nam HJ, Lee D, Yang JS, Kim S, Mair WB, Lee C, Lee SS, Lee SJ. Hwang AB, et al. Proc Natl Acad Sci U S A. 2014 Oct 21;111(42):E4458-67. doi: 10.1073/pnas.1411199111. Epub 2014 Oct 6. Proc Natl Acad Sci U S A. 2014. PMID: 25288734 Free PMC article. - HIF-1 modulates longevity and healthspan in a temperature-dependent manner.
Leiser SF, Begun A, Kaeberlein M. Leiser SF, et al. Aging Cell. 2011 Apr;10(2):318-26. doi: 10.1111/j.1474-9726.2011.00672.x. Epub 2011 Feb 23. Aging Cell. 2011. PMID: 21241450 Free PMC article. - HIF-1 modulates dietary restriction-mediated lifespan extension via IRE-1 in Caenorhabditis elegans.
Chen D, Thomas EL, Kapahi P. Chen D, et al. PLoS Genet. 2009 May;5(5):e1000486. doi: 10.1371/journal.pgen.1000486. Epub 2009 May 22. PLoS Genet. 2009. PMID: 19461873 Free PMC article. - The hypoxia-inducible factor HIF-1 functions as both a positive and negative modulator of aging.
Leiser SF, Kaeberlein M. Leiser SF, et al. Biol Chem. 2010 Oct;391(10):1131-7. doi: 10.1515/BC.2010.123. Biol Chem. 2010. PMID: 20707608 Free PMC article. Review.
Cited by
- The unpredictability of prolonged activation of stress response pathways.
Lamech LT, Haynes CM. Lamech LT, et al. J Cell Biol. 2015 Jun 22;209(6):781-7. doi: 10.1083/jcb.201503107. J Cell Biol. 2015. PMID: 26101215 Free PMC article. - Mitochondrial and cytoplasmic ROS have opposing effects on lifespan.
Schaar CE, Dues DJ, Spielbauer KK, Machiela E, Cooper JF, Senchuk M, Hekimi S, Van Raamsdonk JM. Schaar CE, et al. PLoS Genet. 2015 Feb 11;11(2):e1004972. doi: 10.1371/journal.pgen.1004972. eCollection 2015 Feb. PLoS Genet. 2015. PMID: 25671321 Free PMC article. - Quantitative proteomics analysis of young and elderly skin with DIA mass spectrometry reveals new skin aging-related proteins.
Ma J, Liu M, Wang Y, Xin C, Zhang H, Chen S, Zheng X, Zhang X, Xiao F, Yang S. Ma J, et al. Aging (Albany NY). 2020 Jun 29;12(13):13529-13554. doi: 10.18632/aging.103461. Epub 2020 Jun 29. Aging (Albany NY). 2020. PMID: 32602849 Free PMC article. - Superoxide dismutase is dispensable for normal animal lifespan.
Van Raamsdonk JM, Hekimi S. Van Raamsdonk JM, et al. Proc Natl Acad Sci U S A. 2012 Apr 10;109(15):5785-90. doi: 10.1073/pnas.1116158109. Epub 2012 Mar 26. Proc Natl Acad Sci U S A. 2012. PMID: 22451939 Free PMC article. - Reduction in the levels of CoQ biosynthetic proteins is related to an increase in lifespan without evidence of hepatic mitohormesis.
Rodríguez-Hidalgo M, Luna-Sánchez M, Hidalgo-Gutiérrez A, Barriocanal-Casado E, Mascaraque C, Acuña-Castroviejo D, Rivera M, Escames G, López LC. Rodríguez-Hidalgo M, et al. Sci Rep. 2018 Sep 18;8(1):14013. doi: 10.1038/s41598-018-32190-y. Sci Rep. 2018. PMID: 30228311 Free PMC article.
References
- Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272:1010–1013. - PubMed
- Braeckman BP, Houthoofd K, De Vreese A, Vanfleteren JR. Apparent uncoupling of energy production and consumption in long-lived Clk mutants of Caenorhabditis elegans. Curr Biol. 1999;9:493–496. - PubMed
- Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev. Cell. 2001;1:633–644. - PubMed
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources